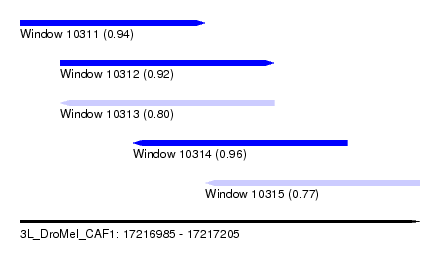
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,216,985 – 17,217,205 |
| Length | 220 |
| Max. P | 0.955686 |

| Location | 17,216,985 – 17,217,087 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -29.08 |
| Energy contribution | -28.88 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17216985 102 + 23771897 ----------------A--CACGUGCUGUGCGGAUUUAUGGCCAAAAGCAAAGGACACGCCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUG ----------------.--.....((((((((((...(((((((((((.....((((.(((..(((......)))..)))))))..))))))))))))).)))).))))........... ( -32.40) >DroSec_CAF1 100518 104 + 1 ----------------ACACACGUGCUGCGCGGAUUUAUGGCCAAAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUG ----------------......((..((((((((....((((((((((.....(((((((........))).........))))..))))))))))))))))))...))........... ( -30.10) >DroSim_CAF1 93754 104 + 1 ----------------ACACACGUGCUGCGCGGAUUUAUGGCCAAAAGCAAAGGACACGGCACGUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUG ----------------......((..((((((((....(((((((((....((...(((((..(((......)))...)))))..)))))))))))))))))))...))........... ( -30.40) >DroEre_CAF1 103847 120 + 1 GCCAACCAGCACCAGCACACACGUGCUGCGCGGAUUUAUGGCCAGAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAAUG ......((((...(((((....)))))(((((((....((((((((((.....(((((((........))).........))))..)))))))))))))))))....))))......... ( -40.20) >DroYak_CAF1 106276 114 + 1 GCCAACCG------GCACACACGUGCUGCGCGGAUUUAUGGCCAGAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAAUG ((.....(------((((....)))))(((((((....((((((((((.....(((((((........))).........))))..)))))))))))))))))....))........... ( -35.60) >consensus ________________ACACACGUGCUGCGCGGAUUUAUGGCCAAAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUG ......................((..((((((((....((((((((((.....(((((((........))).........))))..))))))))))))))))))...))........... (-29.08 = -28.88 + -0.20)


| Location | 17,217,007 – 17,217,125 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -27.28 |
| Energy contribution | -26.80 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17217007 118 + 23771897 GCCAAAAGCAAAGGACACGCCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUGAAGUUAAAAGUGCGUGUUU-UUU-CCGCUUUUCAUUAUCA ...(((((((((((((((((.((.....((.......))...((((((((.(((.....((......)).)))))))))))........))))))))))-)))-..))))))........ ( -32.80) >DroSec_CAF1 100542 119 + 1 GCCAAAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUGAAGUUAAAAGUGCGUGUUU-UUUGCCGCUUUUCAUUAUCA .......((((((((((((.(((.....((.......))...((((((((.(((.....((......)).)))))))))))........))))))))))-)))))............... ( -34.50) >DroSim_CAF1 93778 120 + 1 GCCAAAAGCAAAGGACACGGCACGUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUGAAGUUAAAAGUGCGUGUUUUUUUGCCGCUUUUCAUUAUCA ...((((((((((((((((.(((.....((.......))...((((((((.(((.....((......)).)))))))))))........)))))))))))))....))))))........ ( -32.00) >DroEre_CAF1 103887 117 + 1 GCCAGAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAAUGAAGUUAAAAGUGCGUGUUU-UUU-CCGCUUUUCAUU-CCA ...((((((((((((((((.(((.....((.......))...(((.((((((((.....((......)).)))))))))))........))))))))))-)))-..))))))....-... ( -30.60) >DroYak_CAF1 106310 117 + 1 GCCAGAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAAUGAAGUUAAAAGUGCGUGUUU-UUU-CCGCUUUUCAUU-UCA ...((((((((((((((((.(((.....((.......))...(((.((((((((.....((......)).)))))))))))........))))))))))-)))-..))))))....-... ( -30.60) >consensus GCCAAAAGCAAAGGACACGGCACAUUAACCGAAAUUAGGCUGUCACUUUUUGGCCAUUCGCGCAUUAGCUGCCAAAAGUGAAGUUAAAAGUGCGUGUUU_UUU_CCGCUUUUCAUUAUCA ...((((((...(((((((.(((.....((.......))...(((.((((((((.....((......)).)))))))))))........)))))))))).......))))))........ (-27.28 = -26.80 + -0.48)

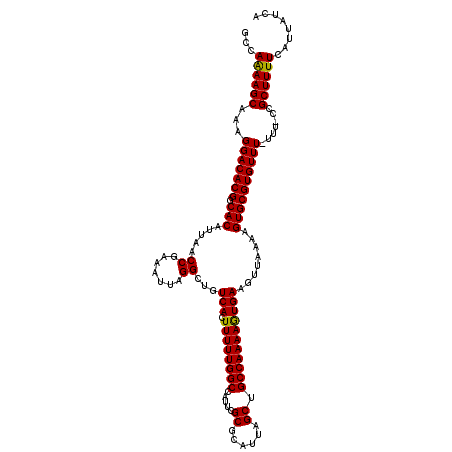
| Location | 17,217,007 – 17,217,125 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -29.68 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17217007 118 - 23771897 UGAUAAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACAGCCUAAUUUCGGUUAAUGUGGCGUGUCCUUUGCUUUUGGC ......(.((((((((-(..-..((((((....(((((((((((((((((.((......)).....))).))))))))............))))))....))))))..))))))))).). ( -34.20) >DroSec_CAF1 100542 119 - 1 UGAUAAUGAAAAGCGGCAAA-AAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACAGCCUAAUUUCGGUUAAUGUGCCGUGUCCUUUGCUUUUGGC ..............((((((-..((((((((..(((((((((((((((((.((......)).....))).))))))))............)))))).))).)))))..))))))...... ( -33.30) >DroSim_CAF1 93778 120 - 1 UGAUAAUGAAAAGCGGCAAAAAAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACAGCCUAAUUUCGGUUAACGUGCCGUGUCCUUUGCUUUUGGC ..............((((((...((((((((........(((((((((((.((......)).....))).)))))))).((((.......))))...))).)))))..))))))...... ( -32.00) >DroEre_CAF1 103887 117 - 1 UGG-AAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUUCAUUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACAGCCUAAUUUCGGUUAAUGUGCCGUGUCCUUUGCUUCUGGC ...-......((((((-(..-..((((((((..(((((((((((((((((.((......)).....))).))))))))............)))))).))).)))))..)))))))..... ( -29.00) >DroYak_CAF1 106310 117 - 1 UGA-AAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUUCAUUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACAGCCUAAUUUCGGUUAAUGUGCCGUGUCCUUUGCUUCUGGC ...-......((((((-(..-..((((((((..(((((((((((((((((.((......)).....))).))))))))............)))))).))).)))))..)))))))..... ( -29.00) >consensus UGAUAAUGAAAAGCGG_AAA_AAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACAGCCUAAUUUCGGUUAAUGUGCCGUGUCCUUUGCUUUUGGC ......(.((((((((.......((((((((........(((((((((((.((......)).....))).)))))))).((((.......))))...))).)))))...)))))))).). (-29.68 = -29.84 + 0.16)

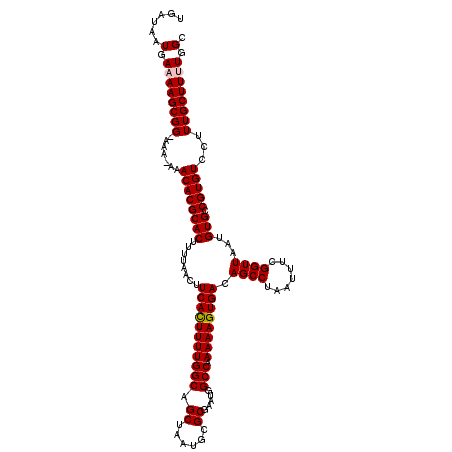
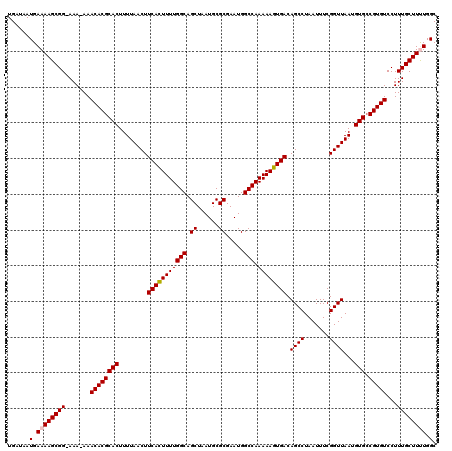
| Location | 17,217,047 – 17,217,165 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17217047 118 - 23771897 CAUGCUUCGCUGCGCGCCAAAAAGUAGACAACAAAAGCCUUGAUAAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACA ......(((((....((((............((((((...(((...(((((.(((.-...-.....)))..)))))...)))))))))((.((....))))...)))).....))))).. ( -24.10) >DroSec_CAF1 100582 119 - 1 CACGCUUCGCUGCGCGGGAAAAAGUAGGCAACAAAAGCCUUGAUAAUGAAAAGCGGCAAA-AAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACA (.(((......))).)(..((((((((((.......))))............(((.....-.....)))))))))..).(((((((((((.((......)).....))).)))))))).. ( -30.20) >DroSim_CAF1 93818 120 - 1 CACGCUUCGCUGCGCGGGAAAAAGUAGGCAACAAAAGCCUUGAUAAUGAAAAGCGGCAAAAAAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACA (.(((......))).)(..((((((((((.......))))............(((...........)))))))))..).(((((((((((.((......)).....))).)))))))).. ( -30.10) >DroEre_CAF1 103927 117 - 1 AACGCUUCGCUGCGCGCCAAAAAGUAGGCAACGAAAGCCUUGG-AAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUUCAUUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACA ..(((((.(((.(((((((((((..((((.......))))(((-(.(((((.(((.-...-.....)))..)))))..)))))))))))).((....)))))...)))....)))))... ( -31.60) >DroYak_CAF1 106350 117 - 1 AACGCUUCGCUGCGCGCCAAAAAGGAGGCAACAAAAGCCUUGA-AAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUUCAUUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACA ..(((((.(((.(((((((((((.(((((.......)))))((-(.(((((.(((.-...-.....)))..)))))..))).)))))))).((....)))))...)))....)))))... ( -33.50) >consensus CACGCUUCGCUGCGCGCCAAAAAGUAGGCAACAAAAGCCUUGAUAAUGAAAAGCGG_AAA_AAACACGCACUUUUAACUUCACUUUUGGCAGCUAAUGCGCGAAUGGCCAAAAAGUGACA ..(((......))).....((((((((((.......))))............(((...........)))))))))....(((((((((((.((......)).....))).)))))))).. (-25.52 = -25.52 + -0.00)

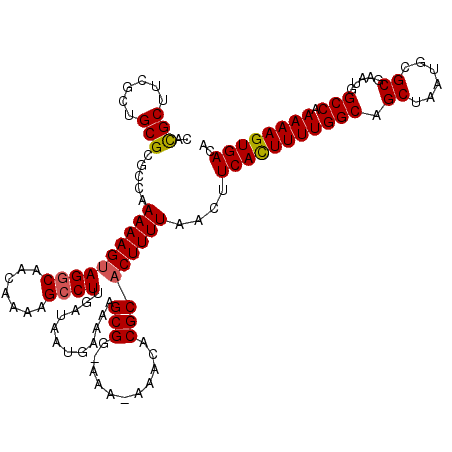
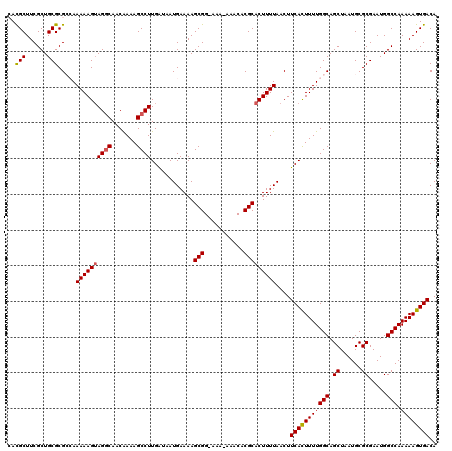
| Location | 17,217,087 – 17,217,205 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17217087 118 - 23771897 AAAUACAACUCAUAUCCUUGCCAAUGACUUCAAAGCAGCGCAUGCUUCGCUGCGCGCCAAAAAGUAGACAACAAAAGCCUUGAUAAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUU .........((((((((((.......((((....((.(((((.((...)))))))))....)))).........)))....))).))))...(((.-...-.....)))........... ( -18.39) >DroSec_CAF1 100622 119 - 1 AAAUACACCUCAUAUCCUUGCCAAGGACUUCAAAGCAGCGCACGCUUCGCUGCGCGGGAAAAAGUAGGCAACAAAAGCCUUGAUAAUGAAAAGCGGCAAA-AAACACGCACUUUUAACUU .......((.(...((((.....)))).......((((((.......))))))).))..((((((((((.......))))............(((.....-.....)))))))))..... ( -26.40) >DroSim_CAF1 93858 120 - 1 AAAUACACCUCAUAUCCUUGCCAAGGACUUCAAAGCAGCGCACGCUUCGCUGCGCGGGAAAAAGUAGGCAACAAAAGCCUUGAUAAUGAAAAGCGGCAAAAAAACACGCACUUUUAACUU .......((.(...((((.....)))).......((((((.......))))))).))..((((((((((.......))))............(((...........)))))))))..... ( -26.30) >DroEre_CAF1 103967 107 - 1 ----------CAUUUCCUAGUUAAGGACUUCAAAGCAGUGAACGCUUCGCUGCGCGCCAAAAAGUAGGCAACGAAAGCCUUGG-AAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUU ----------..((((....((((((..(((...(((((((.....)))))))..(((........)))...)))..))))))-...)))).(((.-...-.....)))........... ( -29.20) >DroYak_CAF1 106390 117 - 1 AAACACACCGCAUAUCCUAGCCAAGGACUUCAAAGCAGUGAACGCUUCGCUGCGCGCCAAAAAGGAGGCAACAAAAGCCUUGA-AAUGAAAAGCGG-AAA-AAACACGCACUUUUAACUU ........(((...((((.....)))).......(((((((.....))))))))))...((((((((((.......)))))..-........(((.-...-.....))).)))))..... ( -29.90) >consensus AAAUACACCUCAUAUCCUUGCCAAGGACUUCAAAGCAGCGCACGCUUCGCUGCGCGCCAAAAAGUAGGCAACAAAAGCCUUGAUAAUGAAAAGCGG_AAA_AAACACGCACUUUUAACUU ..............((((.....)))).......((((((.......))))))......((((((((((.......))))............(((...........)))))))))..... (-20.36 = -20.72 + 0.36)

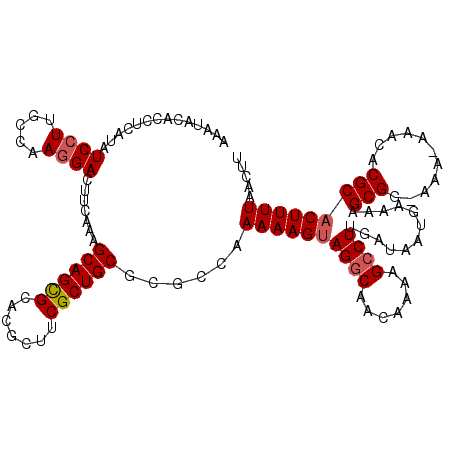
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:31 2006