
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,215,413 – 17,215,644 |
| Length | 231 |
| Max. P | 0.999973 |

| Location | 17,215,413 – 17,215,533 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -28.80 |
| Energy contribution | -30.96 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215413 120 + 23771897 ACGAAUCGAACCCAUAGUUCGCACUUUGGCGAAAUGCGAAAUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCC .(.((((..........((((((.(((...))).))))))(((((.((((((....(((......)))..))))))..((((....))))..........))))))))).)......... ( -31.90) >DroSec_CAF1 99024 120 + 1 UCGAAUCCAACCCAUAGUUCGCACUUUGGCGAAAUGCGGAAUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCC .....((((((((((..(((((......)))))))).)).(((((.((((((....(((......)))..))))))..((((....))))..........)))))..)))))........ ( -35.70) >DroSim_CAF1 92230 120 + 1 UCGAAUCCUACCCAUAGUUCGCACUUUGGCGAAAUGCGGAAUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCC ((.((((...(((((..(((((......)))))))).)).(((((.((((((....(((......)))..))))))..((((....))))..........))))))))).))........ ( -35.00) >DroEre_CAF1 102357 120 + 1 CCGAAUCCAACCCAUGGUUCGCACUUUGGCAAAAUGCGGAAUGGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGCAUGACCCC ((.((((((.....((((((((..(((.((.....)).)))..)).((((((....(((......)))..))))))..((((....))))....))))))..)).))))))......... ( -34.20) >DroYak_CAF1 104633 120 + 1 UCGAAUCCAACCCAUAGUUCGCACUUUGGCGAAAUGCGGAAUGGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGCAUGACCCC (((...(((((((((..(((((......)))))))).)).(((((.((((((....(((......)))..))))))..((((....))))..........)))))..))))..))).... ( -32.20) >consensus UCGAAUCCAACCCAUAGUUCGCACUUUGGCGAAAUGCGGAAUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCC .(.((((...(((((..(((((......)))))))).)).(((((.((((((....(((......)))..))))))..((((....))))..........))))))))).)......... (-28.80 = -30.96 + 2.16)

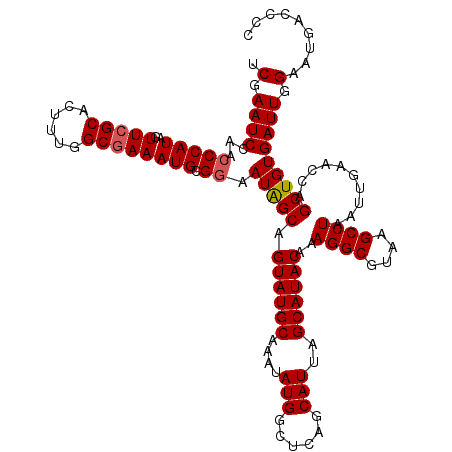
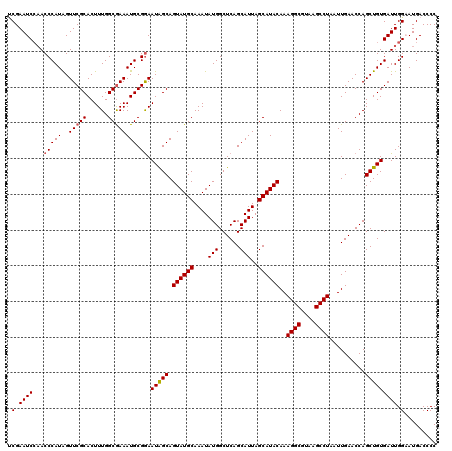
| Location | 17,215,453 – 17,215,564 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -39.16 |
| Energy contribution | -41.16 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.09 |
| SVM RNA-class probability | 0.999973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215453 111 + 23771897 AUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCAAAGUGGGUGGUGUGGGU--------- (((((.((((((....(((......)))..))))))..((((....))))..........))))).............((((.((((((((......)))))))).)))).--------- ( -42.40) >DroSec_CAF1 99064 111 + 1 AUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCCAAGUGGGUGGUGCGGGU--------- (((((.((((((....(((......)))..))))))..((((....))))..........))))).............(((..((((((((......))))))))..))).--------- ( -40.70) >DroSim_CAF1 92270 111 + 1 AUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCCAAGUGGGUGGUGCGGGU--------- (((((.((((((....(((......)))..))))))..((((....))))..........))))).............(((..((((((((......))))))))..))).--------- ( -40.70) >DroEre_CAF1 102397 120 + 1 AUGGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGCAUGACCCCCAAGCCACCCAGAAAAGUGGGUGGUGUGGGUGCUGUGGGU .(.(((((((((...(((((((..((....))...(((((((....))))..)))....)))))))....))))....((((.((((((((......)))))))).)))).))))).).. ( -46.60) >DroYak_CAF1 104673 111 + 1 AUGGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGCAUGACCCCCAAGCCACCCGGAUAAGUGGGUGGUGUGGGU--------- (((.((((.((((....(((.(((((....))......((((....))))...))).)))..)))))))).)))....((((.((((((((......)))))))).)))).--------- ( -42.80) >consensus AUAGCAGUAUGCAAAUAUGGCUCAGCAUUAGCAUACAAAGGCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCAAAGUGGGUGGUGUGGGU_________ (((((.((((((....(((......)))..))))))..((((....))))..........))))).............((((.((((((((......)))))))).)))).......... (-39.16 = -41.16 + 2.00)

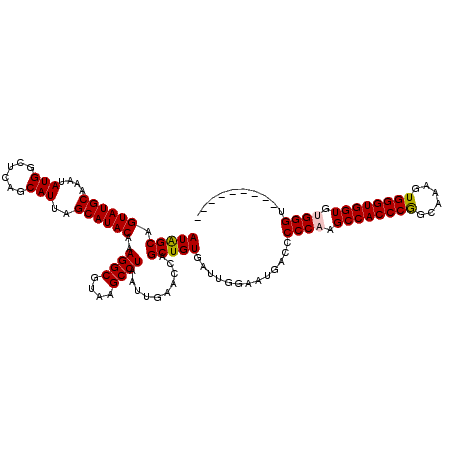
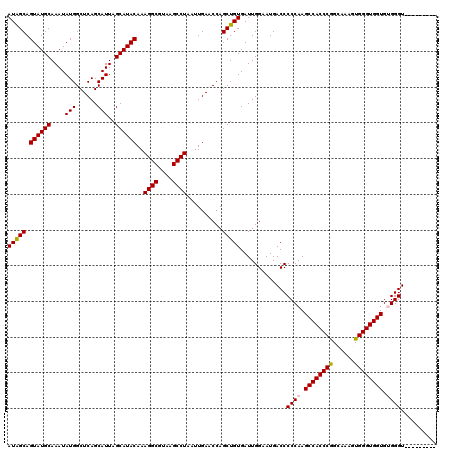
| Location | 17,215,453 – 17,215,564 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -35.84 |
| Energy contribution | -35.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.02 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215453 111 - 23771897 ---------ACCCACACCACCCACUUUGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGCCUUUGUAUGCUAAUGCUGAGCCAUAUUUGCAUACUGCUAU ---------.((((..((((((........))))))..))))((.....)).........(((((((...((((....))))..((((....))))))))))).....((.....))... ( -35.70) >DroSec_CAF1 99064 111 - 1 ---------ACCCGCACCACCCACUUGGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGCCUUUGUAUGCUAAUGCUGAGCCAUAUUUGCAUACUGCUAU ---------.((((..((((((........))))))..))))((.....)).........(((((((...((((....))))..((((....))))))))))).....((.....))... ( -35.00) >DroSim_CAF1 92270 111 - 1 ---------ACCCGCACCACCCACUUGGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGCCUUUGUAUGCUAAUGCUGAGCCAUAUUUGCAUACUGCUAU ---------.((((..((((((........))))))..))))((.....)).........(((((((...((((....))))..((((....))))))))))).....((.....))... ( -35.00) >DroEre_CAF1 102397 120 - 1 ACCCACAGCACCCACACCACCCACUUUUCUGGGUGGCUUGGGGGUCAUGCCAAUCACAGCUGGUUCAAUUAGGCUUACGCCUUUGUAUGCUAAUGCUGAGCCAUAUUUGCAUACUGCCAU .....((((.((((..(((((((......)))))))..))))((.....)).......))))........((((....))))..((((((.((((........)))).))))))...... ( -40.40) >DroYak_CAF1 104673 111 - 1 ---------ACCCACACCACCCACUUAUCCGGGUGGCUUGGGGGUCAUGCCAAUCACAGCUGGUUCAAUUAGGCUUACGCCUUUGUAUGCUAAUGCUGAGCCAUAUUUGCAUACUGCCAU ---------.((((..((((((........))))))..))))(((.((((.(((......(((((((...((((....))))..((((....))))))))))).))).))))...))).. ( -38.60) >consensus _________ACCCACACCACCCACUUGGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGCCUUUGUAUGCUAAUGCUGAGCCAUAUUUGCAUACUGCUAU ..........((((..((((((........))))))..))))((.....)).........(((((((...((((....))))..((((....))))))))))).....((.....))... (-35.84 = -35.60 + -0.24)

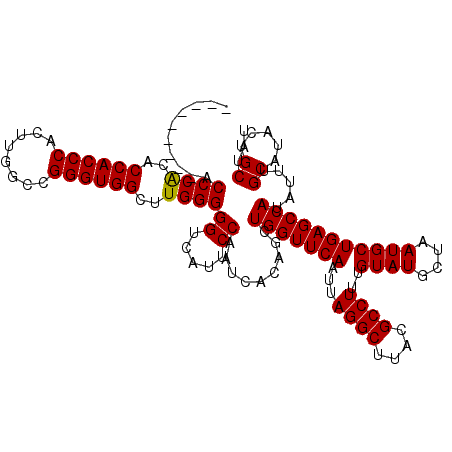

| Location | 17,215,493 – 17,215,604 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215493 111 + 23771897 GCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCAAAGUGGGUGGUGUGGGU---------GGUGCGGAUGGUGCGGCUUCAUCGUUGGGGGCUGACCCAC ((....)).........(((((.((((........(((((((.((((((((......)))))))).)))).---------)))((.(......).)).)))).)))))(((....))).. ( -44.90) >DroSec_CAF1 99104 102 + 1 GCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCCAAGUGGGUGGUGCGGGU------------------GGUUCGGGUUGAUCGUUGGGGGCUGACCCAC (((..(((((....((((((.((((..((((........))))((((((((......)))))))))))).)------------------))))))))))...)))((((......)))). ( -43.60) >DroSim_CAF1 92310 102 + 1 GCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCCAAGUGGGUGGUGCGGGU------------------GGUGCGGGUUCAUCGUUGGGGGCUGACCCAC ((....))(((((((((((.((.....((((........))))((((((((((((......)))).)))))------------------))))).))))))..)))))(((....))).. ( -43.20) >DroEre_CAF1 102437 119 + 1 GCGUAAGCCUAAUUGAACCAGCUGUGAUUGGCAUGACCCCCAAGCCACCCAGAAAAGUGGGUGGUGUGGGUGCUGUGGGUGGUGUGGGUGGUGUGGGUUUUUCGU-GGGGGCUGACCCAC (((.(((((((.....((((.((.......((((.(((((((.((((((((......)))))))).))).......)))).)))))).)))).)))))))..)))-(((......))).. ( -48.62) >DroYak_CAF1 104713 98 + 1 GCGUAAGCCUAAUUGAACCAGCUGUGAUUGGCAUGACCCCCAAGCCACCCGGAUAAGUGGGUGGUGUGGGU---------GGUGCGGGUGGUGUGGGUU-------------UGACCCAC ((....))........((((.((((..........(((((((.((((((((......)))))))).)))).---------))))))).))))(((((..-------------...))))) ( -44.20) >consensus GCGUAAGCCUAAUUGAACCAGCUGUGAUUGGAAUGACCCCCAAGCCACCCGGCAAAGUGGGUGGUGUGGGU_________GGUG_GG_UGGUGCGGGUUCAUCGUUGGGGGCUGACCCAC ......(((........((((......)))).......((((.((((((((......)))))))).))))...................)))..(((((..((......))..))))).. (-30.32 = -30.86 + 0.54)


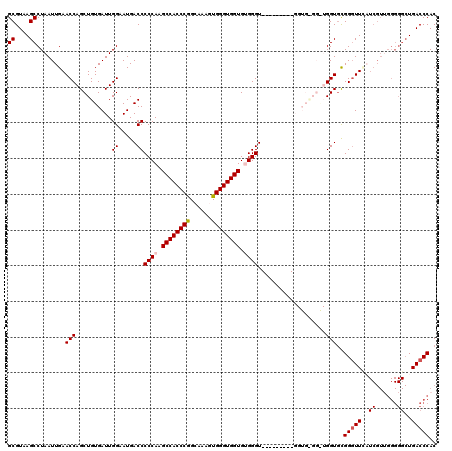
| Location | 17,215,493 – 17,215,604 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.76 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215493 111 - 23771897 GUGGGUCAGCCCCCAACGAUGAAGCCGCACCAUCCGCACC---------ACCCACACCACCCACUUUGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGC (((((((.((((((((.((((.........))))......---------.......((((((........)))))).))))))))....(((........)))........))))))).. ( -36.70) >DroSec_CAF1 99104 102 - 1 GUGGGUCAGCCCCCAACGAUCAACCCGAACC------------------ACCCGCACCACCCACUUGGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGC (((((((.((((((((((.......))..((------------------(((((..(((......))).))))))).))))))))....(((........)))........))))))).. ( -39.00) >DroSim_CAF1 92310 102 - 1 GUGGGUCAGCCCCCAACGAUGAACCCGCACC------------------ACCCGCACCACCCACUUGGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGC (((((((.((((((((((.......))..((------------------(((((..(((......))).))))))).))))))))....(((........)))........))))))).. ( -38.30) >DroEre_CAF1 102437 119 - 1 GUGGGUCAGCCCCC-ACGAAAAACCCACACCACCCACACCACCCACAGCACCCACACCACCCACUUUUCUGGGUGGCUUGGGGGUCAUGCCAAUCACAGCUGGUUCAAUUAGGCUUACGC (((((......)))-))............................((((.((((..(((((((......)))))))..))))((.....)).......)))).................. ( -36.50) >DroYak_CAF1 104713 98 - 1 GUGGGUCA-------------AACCCACACCACCCGCACC---------ACCCACACCACCCACUUAUCCGGGUGGCUUGGGGGUCAUGCCAAUCACAGCUGGUUCAAUUAGGCUUACGC (((((...-------------..))))).((....(.(((---------.((((..((((((........))))))..))))))))..((((........)))).......))....... ( -35.80) >consensus GUGGGUCAGCCCCCAACGAUGAACCCGCACCA_CC_CACC_________ACCCACACCACCCACUUGGCCGGGUGGCUUGGGGGUCAUUCCAAUCACAGCUGGUUCAAUUAGGCUUACGC .((..(((((........................................((((..((((((........))))))..))))((.....)).......)))))..))............. (-29.60 = -29.76 + 0.16)


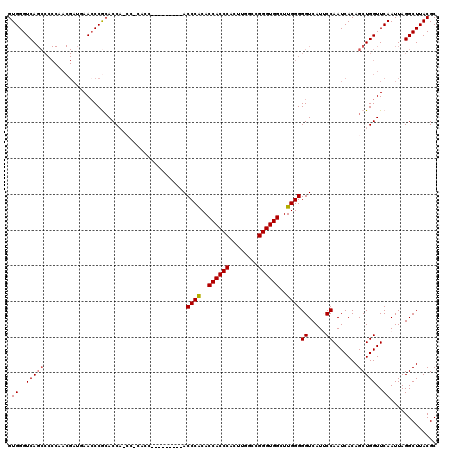
| Location | 17,215,533 – 17,215,644 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215533 111 - 23771897 UUAAUUGCACCACCGUCACCACUAAUUCCCCGCCCGCAUUGUGGGUCAGCCCCCAACGAUGAAGCCGCACCAUCCGCACC---------ACCCACACCACCCACUUUGCCGGGUGGCUUG .............................(((((((((..((((((..((...((....))..)).((.......))...---------.........))))))..)).))))))).... ( -26.80) >DroSec_CAF1 99144 102 - 1 UUAAUUGCACCACCGUCACCACUAAUUCCCCGCCCGCAUUGUGGGUCAGCCCCCAACGAUCAACCCGAACC------------------ACCCGCACCACCCACUUGGCCGGGUGGCUUG .............................((((((((...((((((..((......((.......))....------------------....))...))))))...).))))))).... ( -23.94) >DroSim_CAF1 92350 102 - 1 UUAAUUGCACCACCGUCACCACUAAUUCCCCGCCCGCAUUGUGGGUCAGCCCCCAACGAUGAACCCGCACC------------------ACCCGCACCACCCACUUGGCCGGGUGGCUUG ...............................((...(((((((((......))).)))))).....)).((------------------(((((..(((......))).))))))).... ( -27.30) >DroEre_CAF1 102477 119 - 1 UUAAUUGCACCGCCGUCACCACUAAUUCCCCGCCCGCAUUGUGGGUCAGCCCCC-ACGAAAAACCCACACCACCCACACCACCCACAGCACCCACACCACCCACUUUUCUGGGUGGCUUG .....(((...((.(.(..............).).)).(((((((......)))-))))............................)))......(((((((......))))))).... ( -24.04) >DroYak_CAF1 104753 98 - 1 UUAAUUGCACCACCGUCACCACUAAUUCCCCGCCCGCAUUGUGGGUCA-------------AACCCACACCACCCGCACC---------ACCCACACCACCCACUUAUCCGGGUGGCUUG .....(((.......................(....)..((((((...-------------..))))))......)))..---------.......((((((........)))))).... ( -22.40) >consensus UUAAUUGCACCACCGUCACCACUAAUUCCCCGCCCGCAUUGUGGGUCAGCCCCCAACGAUGAACCCGCACCA_CC_CACC_________ACCCACACCACCCACUUGGCCGGGUGGCUUG .............................(((((((....((((((....................................................)))))).....))))))).... (-19.44 = -19.28 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:24 2006