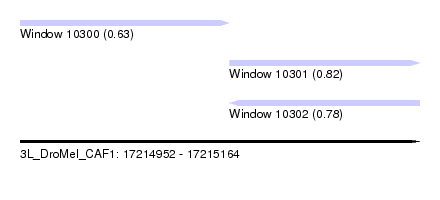
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,214,952 – 17,215,164 |
| Length | 212 |
| Max. P | 0.817053 |

| Location | 17,214,952 – 17,215,063 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
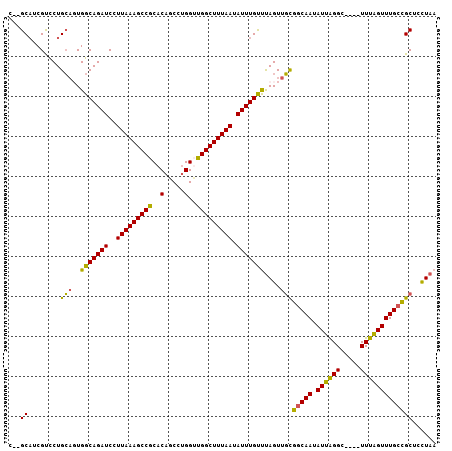
>3L_DroMel_CAF1 17214952 111 + 23771897 CUCGCUACCCCACCACGCCCCUGCC----GUCCUGU-----AACUCAUGCUGUGAAAUUAUCCUCUGCAUGGCAAUCAUAUCAAAUCAAAUUUAAAGUGCCCCUCGCCAGGCACAGUGGG ........(((((........((((----((.....-----.))..((((...((........)).))))))))......................(((((........))))).))))) ( -23.80) >DroSec_CAF1 98622 103 + 1 ------------CCACUCCCCUGCCAUCCAUCCUGU-----AACUCAUGCUGUGAAAUUAUCCUCUGCAUGGCAAUCAUAUCAAAUCAAAUUUAAAGUGCCCCUCGCCCGGCACAGUGGG ------------(((((....((((((.((....((-----((.((((...))))..))))....)).))))))......................(((((........)))))))))). ( -23.70) >DroSim_CAF1 91826 103 + 1 ------------CCACUCCCCUGCCAUCCGUCCUGU-----AACUCAUGCUGUGAAAUUAUCCUCUGCAUGGCAAUCAUAUCAAAUCAAAUUUAAAGUGCCCCUCGCCAGGCACAGUGGG ------------(((((....((((((.((....((-----((.((((...))))..))))....)).))))))......................(((((........)))))))))). ( -23.10) >DroEre_CAF1 101950 97 + 1 --------------CA-UCCCC---ACCCACCCGGU-----AACUCAUGCUGUGAAAUUAUCCGCUGCAUGGCAAUCAUAUCAAAUCAAAUUUAAAGUGCCCCUCGCCAGGCACAGUGGG --------------..-.....---.(((((..(((-----...((((((.(((........))).))))))..)))...................(((((........))))).))))) ( -26.90) >DroYak_CAF1 104186 101 + 1 --------------G--CCCCU---CCCCCCUCGCUGCCUGCACUCAUGCUGUGAAAUUAUCCGCUGCAUGGCAAUCAUAUCAAAUCAAAUUUAAAGUGCCCCUCGCCAGGCACAGUGGA --------------.--.....---......(((((...(((...(((((.(((........))).))))))))......................(((((........)))))))))). ( -22.90) >consensus ____________CCAC_CCCCUGCCACCCAUCCUGU_____AACUCAUGCUGUGAAAUUAUCCUCUGCAUGGCAAUCAUAUCAAAUCAAAUUUAAAGUGCCCCUCGCCAGGCACAGUGGG ..................(((..........................(((((((.............)))))))......................(((((........)))))...))) (-16.56 = -16.76 + 0.20)

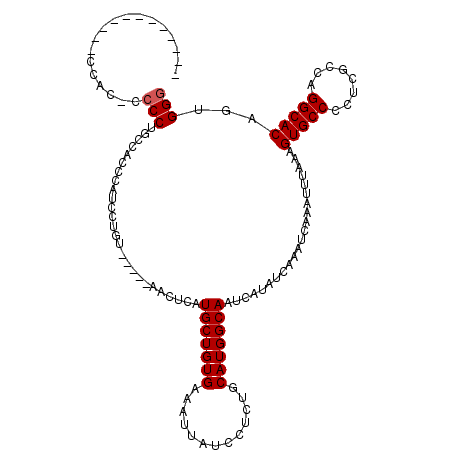
| Location | 17,215,063 – 17,215,164 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -25.64 |
| Energy contribution | -24.20 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
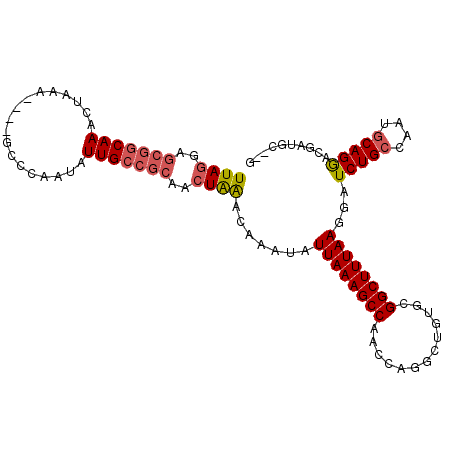
>3L_DroMel_CAF1 17215063 101 + 23771897 C--GCAUCGUCCUGCCGUGGCAGAUCCUUAAAGCCACACAGCCUGGCUGGCUUUAAUAUUUGUUUAGUUGCGGCAAUAUUAGGC----UUUAGUUUGCCGCUUCUAA .--((.......(((((..((((((..(((((((((..(.....)..))))))))).....)))).))..)))))......(((----........)))))...... ( -32.10) >DroSec_CAF1 98725 101 + 1 C--GCAUCGUCCUGCAUUGGCAGAUCCUUAAAGCCGCACAGCCUGGUUGGCUUUAAUAUUUGUUUAGUUGCGGCAAUAUUGGGC----UUUAGUUUGCCGCUCCUAA .--(((......)))...(((((((..(((((((((.((......))))))))))).)))))))(((..(((((((.((((...----..)))))))))))..))). ( -30.30) >DroSim_CAF1 91929 101 + 1 C--GCAUCGUCCUGCAUUGGCAGAUCCUUAAAGCCGCACAGCCUGGUUGGCUUUAAUAUUUGUUUAGUUGCGGCAAUAUUGGGC----UUUAGUUUGCCGCUCCUAA .--(((......)))...(((((((..(((((((((.((......))))))))))).)))))))(((..(((((((.((((...----..)))))))))))..))). ( -30.30) >DroYak_CAF1 104287 106 + 1 CUGGCUUCGCUCUGGAGUGGCAGAUCCUUAAAGCCGC-CAGCCAGGAUGGCUUUAAUAUUUGCCCAGUUCUUCCAAUAUUAGGUUUAGUUUGGUUUGCGGCGCUUAA ..(((.((((...(((.((((((((..((((((((((-(.....)).))))))))).))))).))).)))..((((.((((....))))))))...)))).)))... ( -32.60) >consensus C__GCAUCGUCCUGCAGUGGCAGAUCCUUAAAGCCGCACAGCCUGGUUGGCUUUAAUAUUUGUUUAGUUGCGGCAAUAUUAGGC____UUUAGUUUGCCGCUCCUAA ...((........(((..(((((((..(((((((((..(.....)..))))))))).)))))))....)))(((((.((((((.....)))))))))))))...... (-25.64 = -24.20 + -1.44)

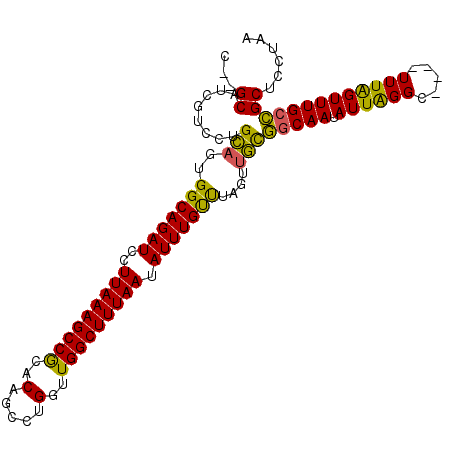
| Location | 17,215,063 – 17,215,164 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -20.48 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17215063 101 - 23771897 UUAGAAGCGGCAAACUAAA----GCCUAAUAUUGCCGCAACUAAACAAAUAUUAAAGCCAGCCAGGCUGUGUGGCUUUAAGGAUCUGCCACGGCAGGACGAUGC--G ((((..(((((((......----........)))))))..)))).......(((((((((..(.....)..)))))))))...(((((....))))).......--. ( -30.94) >DroSec_CAF1 98725 101 - 1 UUAGGAGCGGCAAACUAAA----GCCCAAUAUUGCCGCAACUAAACAAAUAUUAAAGCCAACCAGGCUGUGCGGCUUUAAGGAUCUGCCAAUGCAGGACGAUGC--G ((((..(((((((......----........)))))))..)))).......((((((((.((......))..))))))))(.(((..((......))..))).)--. ( -28.74) >DroSim_CAF1 91929 101 - 1 UUAGGAGCGGCAAACUAAA----GCCCAAUAUUGCCGCAACUAAACAAAUAUUAAAGCCAACCAGGCUGUGCGGCUUUAAGGAUCUGCCAAUGCAGGACGAUGC--G ((((..(((((((......----........)))))))..)))).......((((((((.((......))..))))))))(.(((..((......))..))).)--. ( -28.74) >DroYak_CAF1 104287 106 - 1 UUAAGCGCCGCAAACCAAACUAAACCUAAUAUUGGAAGAACUGGGCAAAUAUUAAAGCCAUCCUGGCUG-GCGGCUUUAAGGAUCUGCCACUCCAGAGCGAAGCCAG ....((..(((...((((.............)))).....(((((((.((.((((((((..((.....)-).))))))))..)).)))....)))).)))..))... ( -27.02) >consensus UUAGGAGCGGCAAACUAAA____GCCCAAUAUUGCCGCAACUAAACAAAUAUUAAAGCCAACCAGGCUGUGCGGCUUUAAGGAUCUGCCAAUGCAGGACGAUGC__G ((((..(((((((..................)))))))..)))).......((((((((.............))))))))...(((((....))))).......... (-20.48 = -21.42 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:19 2006