
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,213,836 – 17,214,378 |
| Length | 542 |
| Max. P | 0.999910 |

| Location | 17,213,836 – 17,213,948 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -18.98 |
| Energy contribution | -20.14 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213836 112 - 23771897 AAUAGCCAUAAAUGCCUUAAAUGCUCGCCGCAGACCGUUUCAAAUUGAAGGCAACCUUAAAUAAUUGACGCUAUCAAGAGGACGAAAAAUUGUCCGAAAUGUCU--------AGCAAUUU ............(((.......((.....))((((.(((((...((((.(((.................))).))))..((((((....)))))))))))))))--------.))).... ( -22.63) >DroSec_CAF1 97550 120 - 1 AAUAGCCAUAAAUGCCUUAAAUGCUCGGCGCAGACCGUUUCAAAUUGAAGGCAACCUUAAAUAAUUGAGGCUAUCAAGAGGACGAAAAAUUGUCCGAAAUGGCAACAAGCAAAGCAUUUU ..................(((((((..((.....(((((((...((((.(((...(((((....)))))))).))))..((((((....)))))))))))))......))..))))))). ( -32.60) >DroSim_CAF1 90758 120 - 1 AAUAGCCACAAAUGCCUUAAAUGCUCGGCGCAGACCGUUUCAAAUUGAAGGCAACCUUAAAUAAUUGAGGCUAUCAAGAGGACGAAAAAUUGUCCGAAAUGGCAACAAGCAAAGCAAUUU .....................((((..((.....(((((((...((((.(((...(((((....)))))))).))))..((((((....)))))))))))))......))..)))).... ( -30.50) >DroEre_CAF1 100924 112 - 1 AAUAGCCAUAACUGCCUUAAAUGGUCGUAGCAGACCGUUUCAAAUUGGAGGAAACUUUAAAUAAUUGAGGCUAUCAAGAGUACCAAAAAUUGUCCAAAAUGGCC--------AGCAAUUU ....(((((....(((((((((((((......))))))......((((((....))))))....)))))))........(.((........)).)...))))).--------........ ( -27.80) >DroYak_CAF1 103124 112 - 1 AAUAGCCAUAAAUGCCUUAAAUGCUCCAAGCAGACCGUUUUAAAUUGGAGCCAACUUUAAAUAAUUGAGGCCAUCAAGAGGACGAAAAAUUGCCCAAAAUGGCC--------AGCAAUUU ............(((.......(((((((...............))))))).................((((((...(.((.(((....))))))...))))))--------.))).... ( -23.86) >consensus AAUAGCCAUAAAUGCCUUAAAUGCUCGGCGCAGACCGUUUCAAAUUGAAGGCAACCUUAAAUAAUUGAGGCUAUCAAGAGGACGAAAAAUUGUCCGAAAUGGCA________AGCAAUUU ....(((((....(((((((.(((.....)))............((((.(....).))))....)))))))........((((((....))))))...)))))................. (-18.98 = -20.14 + 1.16)


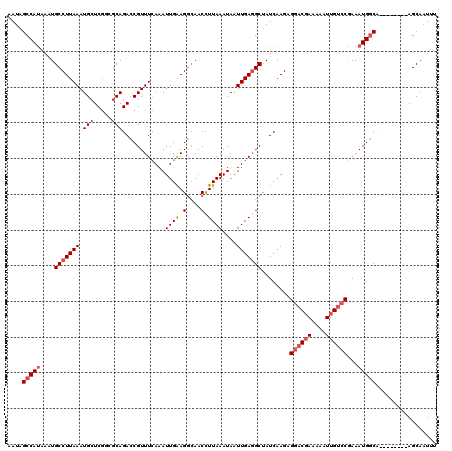
| Location | 17,213,908 – 17,214,028 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213908 120 + 23771897 AAACGGUCUGCGGCGAGCAUUUAAGGCAUUUAUGGCUAUUAAAGUUGAAAAAUGUUUCACUUGCAGUUGAUUUGCAUAAAUAUUUAAACUUGUUUAGCUUGCAUAAGCAUUUUUGCCUCU ........(((.....)))....(((((.....(((((...(((((...((((((((....(((((.....))))).)))))))).)))))...)))))(((....)))....))))).. ( -27.50) >DroSec_CAF1 97630 120 + 1 AAACGGUCUGCGCCGAGCAUUUAAGGCAUUUAUGGCUAUUAAAGUUGAAAAAUGUUUCACUUGCAGUUGAUUUGCAUAAAUAUUUAAACUUGUUUAGCUGGCAUAAGCAUUUUUGCCUCU ...((((....))))........(((((.....(((((...(((((...((((((((....(((((.....))))).)))))))).)))))...))))).((....)).....))))).. ( -29.40) >DroSim_CAF1 90838 120 + 1 AAACGGUCUGCGCCGAGCAUUUAAGGCAUUUGUGGCUAUUAAAGUUGAAAAAUGUUUCACUUGCAGUUGAUUUGCAUAAAUAUUUAAACUUGUUUAGCUGGCAUAAGCAUUUUUGCCUCU ...((((....))))........(((((.....(((((...(((((...((((((((....(((((.....))))).)))))))).)))))...))))).((....)).....))))).. ( -29.40) >DroEre_CAF1 100996 120 + 1 AAACGGUCUGCUACGACCAUUUAAGGCAGUUAUGGCUAUUAAAGUUUAAAAAUGUUUCACUUGCAGUUGAUUUGCAUAAAUAUUUAAACUUGUUUAGCUUGCGUAAGCAUUUUUGCCUCU ....((((......)))).....((((((....(((((...((((((((..((((((....(((((.....))))).))))))))))))))...)))))(((....)))...)))))).. ( -34.90) >DroYak_CAF1 103196 120 + 1 AAACGGUCUGCUUGGAGCAUUUAAGGCAUUUAUGGCUAUUAAAGUUUAAAAAUGUUUCACUUGCAGUUGAUUUGCAUAAAUAUUUAAACUUGUUUAGCUUGCAUAAGCAUUUUUGCCUCU ....(.(((....))).).....(((((.....(((((...((((((((..((((((....(((((.....))))).))))))))))))))...)))))(((....)))....))))).. ( -28.30) >consensus AAACGGUCUGCGCCGAGCAUUUAAGGCAUUUAUGGCUAUUAAAGUUGAAAAAUGUUUCACUUGCAGUUGAUUUGCAUAAAUAUUUAAACUUGUUUAGCUUGCAUAAGCAUUUUUGCCUCU ....(.((......)).).....(((((.....(((((...(((((...((((((((....(((((.....))))).)))))))).)))))...))))).((....)).....))))).. (-25.54 = -25.94 + 0.40)

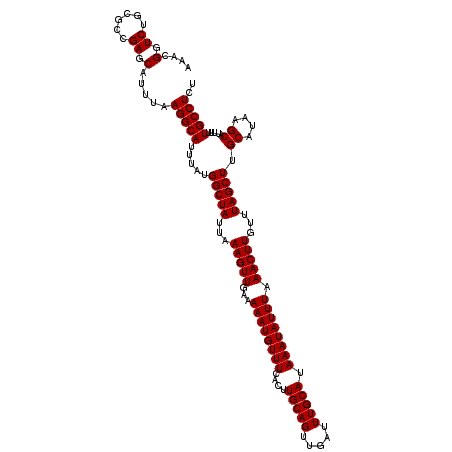
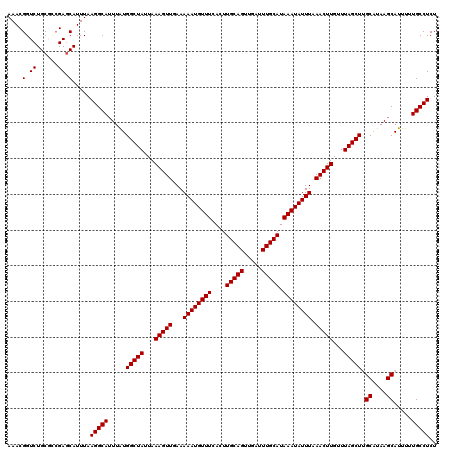
| Location | 17,213,908 – 17,214,028 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213908 120 - 23771897 AGAGGCAAAAAUGCUUAUGCAAGCUAAACAAGUUUAAAUAUUUAUGCAAAUCAACUGCAAGUGAAACAUUUUUCAACUUUAAUAGCCAUAAAUGCCUUAAAUGCUCGCCGCAGACCGUUU .((((((....(((....))).((((...(((((..........((((.......))))...((((....)))))))))...))))......))))))(((((.((......)).))))) ( -22.70) >DroSec_CAF1 97630 120 - 1 AGAGGCAAAAAUGCUUAUGCCAGCUAAACAAGUUUAAAUAUUUAUGCAAAUCAACUGCAAGUGAAACAUUUUUCAACUUUAAUAGCCAUAAAUGCCUUAAAUGCUCGGCGCAGACCGUUU .((((((.....((....))..((((...(((((..........((((.......))))...((((....)))))))))...))))......))))))(((((.((......)).))))) ( -20.80) >DroSim_CAF1 90838 120 - 1 AGAGGCAAAAAUGCUUAUGCCAGCUAAACAAGUUUAAAUAUUUAUGCAAAUCAACUGCAAGUGAAACAUUUUUCAACUUUAAUAGCCACAAAUGCCUUAAAUGCUCGGCGCAGACCGUUU .((((((.....((....))..((((...(((((..........((((.......))))...((((....)))))))))...))))......))))))(((((.((......)).))))) ( -20.80) >DroEre_CAF1 100996 120 - 1 AGAGGCAAAAAUGCUUACGCAAGCUAAACAAGUUUAAAUAUUUAUGCAAAUCAACUGCAAGUGAAACAUUUUUAAACUUUAAUAGCCAUAACUGCCUUAAAUGGUCGUAGCAGACCGUUU .((((((....(((....))).((((...(((((((((..((((((((.......))))..)))).....)))))))))...))))......))))))((((((((......)))))))) ( -31.10) >DroYak_CAF1 103196 120 - 1 AGAGGCAAAAAUGCUUAUGCAAGCUAAACAAGUUUAAAUAUUUAUGCAAAUCAACUGCAAGUGAAACAUUUUUAAACUUUAAUAGCCAUAAAUGCCUUAAAUGCUCCAAGCAGACCGUUU .((((((....(((....))).((((...(((((((((..((((((((.......))))..)))).....)))))))))...))))......))))))...(((.....)))........ ( -25.10) >consensus AGAGGCAAAAAUGCUUAUGCAAGCUAAACAAGUUUAAAUAUUUAUGCAAAUCAACUGCAAGUGAAACAUUUUUCAACUUUAAUAGCCAUAAAUGCCUUAAAUGCUCGGCGCAGACCGUUU .((((((.....((....))..((((...(((((.((((.(((.((((.......))))....))).))))...)))))...))))......))))))(((((.((......)).))))) (-20.54 = -20.54 + -0.00)



| Location | 17,213,988 – 17,214,098 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -22.92 |
| Energy contribution | -22.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213988 110 - 23771897 AAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCA--CAGGAUGCUGGAUUUUCAUGGCAAAAGUCAGAGGCAAAAAUGCUUAUGCAAGCUAAACAAGUUUAAAUA .........((((((((((((--------.........(((....))).--(((....)))........))))....(.((.(((((....))))).))).......))))))))..... ( -23.10) >DroSec_CAF1 97710 112 - 1 AAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCACACAGGAUGCUGGAUUUUCAUGGCAAAAGUCAGAGGCAAAAAUGCUUAUGCCAGCUAAACAAGUUUAAAUA .........((((((((....--------.........(((....))).........(((((.......((((....)))).(((((....)))))...)))))...))))))))..... ( -24.90) >DroSim_CAF1 90918 112 - 1 AAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCACACAGGAUGCUGGAUUUUCAUGGCAAAAGUCAGAGGCAAAAAUGCUUAUGCCAGCUAAACAAGUUUAAAUA .........((((((((....--------.........(((....))).........(((((.......((((....)))).(((((....)))))...)))))...))))))))..... ( -24.90) >DroEre_CAF1 101076 110 - 1 AAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCA--GAGGAUGCUGGAAUUUCAUGGCAAAAGUCAGAGGCAAAAAUGCUUACGCAAGCUAAACAAGUUUAAAUA .........(((((((((((.--------........(((((....((.--..)).))))).....((.((((....)))))))))......((((....))))...))))))))..... ( -22.10) >DroYak_CAF1 103276 118 - 1 AAUCACUUAAAAUUUGUGCCAAACAGCCAAACAAAUACGGCAAAAGCCA--CAGGAUGCUGGAUUUUCAUGGCAAAAGUCAGAGGCAAAAAUGCUUAUGCAAGCUAAACAAGUUUAAAUA .........(((((((((((...((((...........(((....))).--......)))).....((.((((....))))))))).....(((....)))......))))))))..... ( -23.67) >consensus AAUCACUUAAAAUUUGUGCCA________AACAAAUACGGCAAAAGCCA__CAGGAUGCUGGAUUUUCAUGGCAAAAGUCAGAGGCAAAAAUGCUUAUGCAAGCUAAACAAGUUUAAAUA .........(((((((((((.................(((((....((.....)).))))).....((.((((....)))))))))......(((......)))...))))))))..... (-22.92 = -22.92 + -0.00)


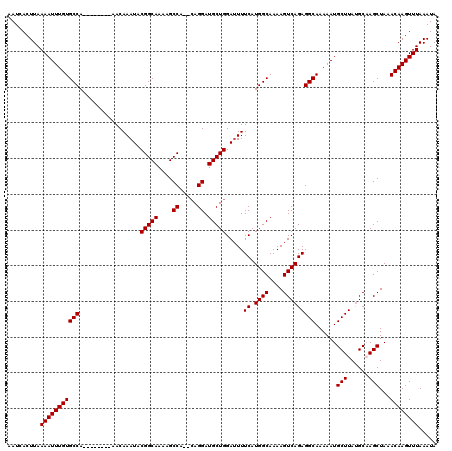
| Location | 17,214,028 – 17,214,138 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -28.68 |
| Energy contribution | -28.64 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214028 110 - 23771897 UUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCA--CAGGAUGCUGGAUUUUCAUGGCAAAAGUC .....((((...((((((...))))))))))((((....(((((.......((((((....--------.))))))..(((....))).--(((....))))))))...))))....... ( -30.60) >DroSec_CAF1 97750 112 - 1 UUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCACACAGGAUGCUGGAUUUUCAUGGCAAAAGUC .....((((...((((((...))))))))))((((....(((((.......((((((....--------.)))))).(((((....((.....)).))))))))))...))))....... ( -31.70) >DroSim_CAF1 90958 112 - 1 UUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCACACAGGAUGCUGGAUUUUCAUGGCAAAAGUC .....((((...((((((...))))))))))((((....(((((.......((((((....--------.)))))).(((((....((.....)).))))))))))...))))....... ( -31.70) >DroEre_CAF1 101116 110 - 1 UUCUCCAUAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGGCAAAAGCCA--GAGGAUGCUGGAAUUUCAUGGCAAAAGUC ....((((....((((((...)))))).....(((((((...((.......((((((....--------.))))))..(((....))).--))..)))))))......))))........ ( -29.40) >DroYak_CAF1 103316 118 - 1 UUCUCCGUAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCAAACAGCCAAACAAAUACGGCAAAAGCCA--CAGGAUGCUGGAUUUUCAUGGCAAAAGUC ...(((((((((.((.(((.((((....).)))..((((((((........))))))))......))).)).))))))(((....))).--..)))(((((........)))))...... ( -31.30) >consensus UUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA________AACAAAUACGGCAAAAGCCA__CAGGAUGCUGGAUUUUCAUGGCAAAAGUC .....((((...((((((...))))))))))((((((((((((........))))))))..................(((((....((.....)).)))))........))))....... (-28.68 = -28.64 + -0.04)

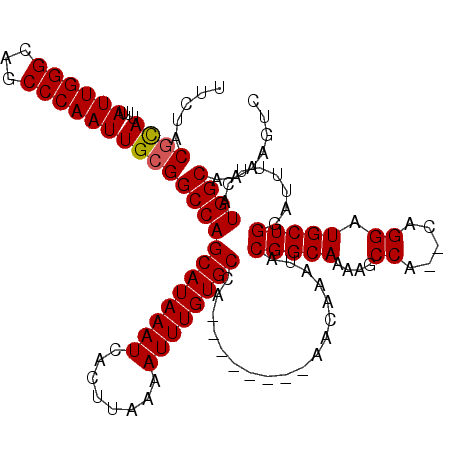

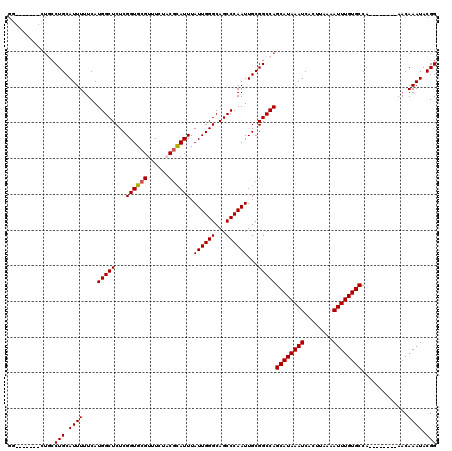
| Location | 17,214,066 – 17,214,178 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -22.94 |
| Energy contribution | -22.90 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214066 112 - 23771897 GGCUCGUUGCUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGG (((.(((.(((((((((..........))....(((((((....)))))))....)))))))......)))))).((((((((........))))))))..--------........... ( -33.30) >DroSec_CAF1 97790 105 - 1 GG-------CUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGG .(-------(((.(((((........((((((((((((((....))))))....)))).))))....))))).))))..............((((((....--------.)))))).... ( -31.20) >DroSim_CAF1 90998 105 - 1 GG-------CUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGG .(-------(((.(((((........((((((((((((((....))))))....)))).))))....))))).))))..............((((((....--------.)))))).... ( -31.20) >DroEre_CAF1 101154 107 - 1 G-----AUGCUCCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUCCAUAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA--------AACAAAUACGG .-----(((((..(((((........(((((((((((.((........)).))))))).))))....)))))..)))))............((((((....--------.)))))).... ( -25.40) >DroYak_CAF1 103354 107 - 1 -------------CUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUCCGUAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCAAACAGCCAAACAAAUACGG -------------..(((((.............)))))......((((((((.((.(((.((((....).)))..((((((((........))))))))......))).)).)))))))) ( -26.52) >consensus GG_______CUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUAAAUCACUUAAAAUUUGUGCCA________AACAAAUACGG .............(((.((((....(((((...((((((......)))))).((((((...))))))...)))))((((((((........)))))))).............)))).))) (-22.94 = -22.90 + -0.04)


| Location | 17,214,098 – 17,214,218 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -41.44 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214098 120 + 23771897 UAUGCUGGCCGCAAUUGGGCUGCCCAAUAAAUGCGUAGAAACGCACCGAGAGCCAUGAAAAAUGCAGGCAGCAACGAGCCGAGCACCGUAUGUUUGGCCAUCUCUCCAUUCGGCCAUGCA ..(((((((((....(((((((((.......(((((....)))))......((.((.....)))).))))))...(.((((((((.....)))))))))......)))..)))))).))) ( -46.10) >DroSec_CAF1 97822 112 + 1 UAUGCUGGCCGCAAUUGGGCUGCCCAAUAAAUGCGUAGAAACGCACCGAGAGCCAUGAAAAAUGCAGGCAG-------CCGAACACCAUCUU-UUGGCCAUCUCUCCAUUCGGCCAUGCA ..(((((((((..((((((...))))))...(((((....)))))..(((((((.((.......))))).(-------(((((........)-)))))..))))......)))))).))) ( -39.10) >DroSim_CAF1 91030 113 + 1 UAUGCUGGCCGCAAUUGGGCUGCCCAAUAAAUGCGUAGAAACGCACCGAGAGCCAUGAAAAAUGCAGGCAG-------CCGAACACCAUCUUUUUGGCCAUCUCGCCAUUCGGCCAUGCA ..(((((((((...((.(((((((.......(((((....)))))......((.((.....)))).)))))-------)).))...........((((......))))..)))))).))) ( -40.60) >DroEre_CAF1 101186 111 + 1 UAUGCUGGCCGCAAUUGGGCUGCCCAAUAAAUAUGGAGAAACGCACCGAGAGCCAUGAAAAAUGCAGGGAGCAU-----CGUGUGCCA--UGUUCGGGUAU--CUCCAUUCGGCCAUGCA ..(((((((((..((((((...))))))....(((((((...((.(((((.(((((((....(((.....))))-----)))).))..--..))))))).)--)))))).)))))).))) ( -43.40) >DroYak_CAF1 103394 102 + 1 UAUGCUGGCCGCAAUUGGGCUGCCCAAUAAAUACGGAGAAACGCACCGAGAGCCAUGAAAAAUGCAG--------------UGUGCCG--UGUUCGGGUAU--CUCCAUUCGGCCAUGCA ..(((((((((.(((.(((.(((((....(((((((....((((..(....).(((.....)))..)--------------))).)))--)))).))))).--))).))))))))).))) ( -38.00) >consensus UAUGCUGGCCGCAAUUGGGCUGCCCAAUAAAUGCGUAGAAACGCACCGAGAGCCAUGAAAAAUGCAGGCAG_______CCGAGCACCAU_UGUUUGGCCAUCUCUCCAUUCGGCCAUGCA ..(((((((((((((((((...))))))...((((......)))).................)))....................(((......)))..............))))).))) (-25.74 = -25.90 + 0.16)



| Location | 17,214,098 – 17,214,218 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -29.94 |
| Energy contribution | -29.42 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214098 120 - 23771897 UGCAUGGCCGAAUGGAGAGAUGGCCAAACAUACGGUGCUCGGCUCGUUGCUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUA (((.((((((....(((((((((((........)))....(((........)))..))))))))..((((((((((((((....))))))....)))).))))......))))))))).. ( -45.30) >DroSec_CAF1 97822 112 - 1 UGCAUGGCCGAAUGGAGAGAUGGCCAA-AAGAUGGUGUUCGG-------CUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUA (((.((((((........((..((((.-....))))..))((-------((((((((..........))....(((((((....)))))))....))))))))......))))))))).. ( -43.20) >DroSim_CAF1 91030 113 - 1 UGCAUGGCCGAAUGGCGAGAUGGCCAAAAAGAUGGUGUUCGG-------CUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUA (((.((((((..((((......))))..............((-------((((((((..........))....(((((((....)))))))....))))))))......))))))))).. ( -43.90) >DroEre_CAF1 101186 111 - 1 UGCAUGGCCGAAUGGAG--AUACCCGAACA--UGGCACACG-----AUGCUCCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUCCAUAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUA (((.((((((.((((((--(.((((((.((--(((.....(-----((((.....)))))..)))))....))))...)).)))))))....((((((...))))))..))))))))).. ( -42.50) >DroYak_CAF1 103394 102 - 1 UGCAUGGCCGAAUGGAG--AUACCCGAACA--CGGCACA--------------CUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUCCGUAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUA (((.((((((.((((((--(.((.((....--))((((.--------------..((..........)).....)))))).)))))))....((((((...))))))..))))))))).. ( -36.30) >consensus UGCAUGGCCGAAUGGAGAGAUGGCCAAACA_AUGGUGCUCGG_______CUGCCUGCAUUUUUCAUGGCUCUCGGUGCGUUUCUACGCAUUUAUUGGGCAGCCCAAUUGCGGCCAGCAUA (((.((((((..(((........))).......(((....................(((.....)))((((..((((((......))))))....)))).)))......))))))))).. (-29.94 = -29.42 + -0.52)

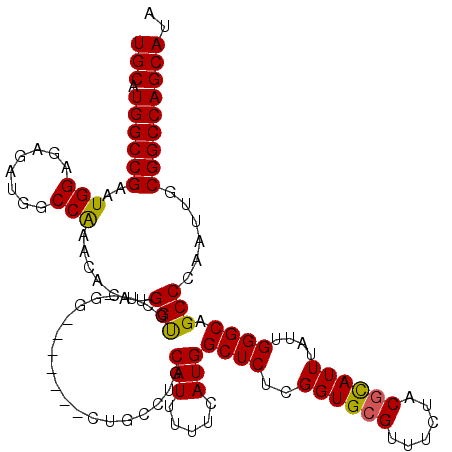
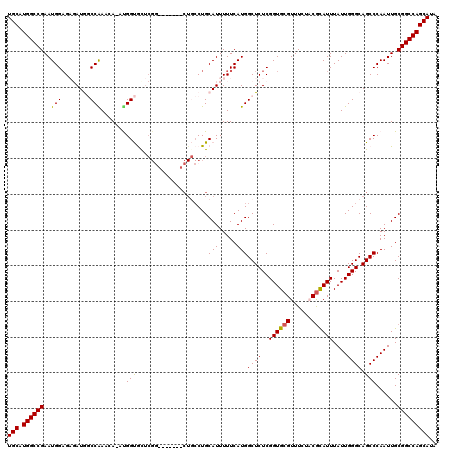
| Location | 17,214,178 – 17,214,286 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214178 108 + 23771897 GAGCACCGUAUGUUUGGCCAUCUCUCCAUUCGGCCAUGCAUAUUUAUUUGCCAACAUUUACUUAUGCUGCUGUGCU--------GUCUCC----UUUUGGCCAAUGGCCGUCGAACUCAA .(((((.(((....(((((............))))).(((((..((..((....))..))..)))))))).)))))--------......----....((((...))))........... ( -28.30) >DroSec_CAF1 97895 104 + 1 GAACACCAUCUU-UUGGCCAUCUCUCCAUUCGGCCAUGCAUAUUUAUUUGCCAACAUUUACUUAUGC---UGUGCU--------GUCUCC----UUGUGGCCAAUGGCCGUCGAACUCAA ((...((((...-.(((((((.........((((((.(((((..((..((....))..))..)))))---)).)))--------).....----..)))))))))))...))........ ( -27.59) >DroSim_CAF1 91103 105 + 1 GAACACCAUCUUUUUGGCCAUCUCGCCAUUCGGCCAUGCAUAUUUAUUUGCCAACAUUUACUUAUGC---UGUGCU--------GUCUCC----UUGUGGCCAAUGGCCGUCGAACUCAA ...............((((((...(((((.((((((.(((((..((..((....))..))..)))))---)).)))--------).....----..)))))..))))))........... ( -29.10) >DroEre_CAF1 101261 104 + 1 GUGUGCCA--UGUUCGGGUAU--CUCCAUUCGGCCAUGCAUAUUUAUUUGCCAACAUUUACUUAUGC---UGCAUUUGCGGCCAGCCGCU----UCUUGGCC-AUGGCC----AACUCAA .((.((((--(....(((...--.)))....(((((.(((((..((..((....))..))..)))))---.......((((....)))).----...)))))-))))))----)...... ( -32.30) >DroYak_CAF1 103461 107 + 1 -UGUGCCG--UGUUCGGGUAU--CUCCAUUCGGCCAUGCAUAUUUAUUUGCCAACAUUUACUUAUGC---UGCAUUUGAGGCAAGUCGUUCUGGUCUCGGCC-AUGGCC----AACUCAA -((.((((--((..((((...--..(((..((((...(((((..((..((....))..))..)))))---(((.......))).))))...))).))))..)-))))))----)...... ( -31.70) >consensus GAGCACCAU_UGUUUGGCCAUCUCUCCAUUCGGCCAUGCAUAUUUAUUUGCCAACAUUUACUUAUGC___UGUGCU________GUCUCC____UUUUGGCCAAUGGCCGUCGAACUCAA .....(((......)))..............(((((((((((..((..((....))..))..))))).................(((...........)))..))))))........... (-17.52 = -17.28 + -0.24)


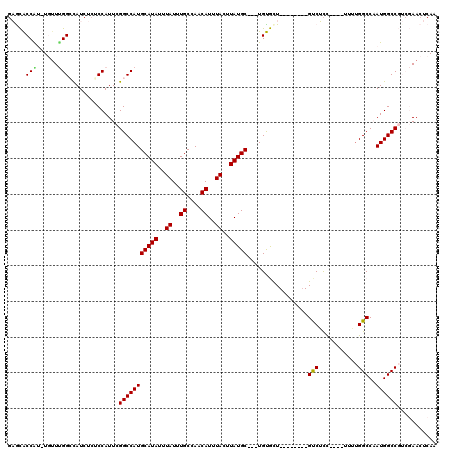
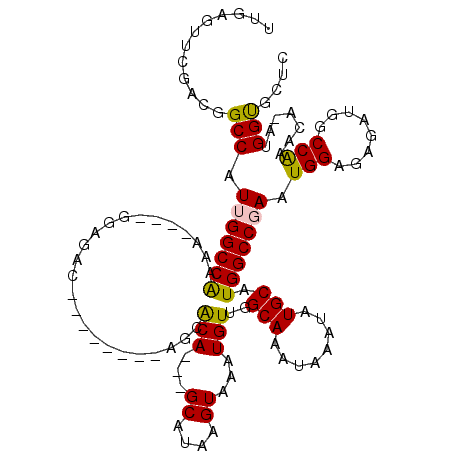
| Location | 17,214,178 – 17,214,286 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -19.20 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214178 108 - 23771897 UUGAGUUCGACGGCCAUUGGCCAAAA----GGAGAC--------AGCACAGCAGCAUAAGUAAAUGUUGGCAAAUAAAUAUGCAUGGCCGAAUGGAGAGAUGGCCAAACAUACGGUGCUC ...........((((...))))....----(....)--------(((((..((((((......)))))).........((((..((((((..(....)..))))))..))))..))))). ( -29.80) >DroSec_CAF1 97895 104 - 1 UUGAGUUCGACGGCCAUUGGCCACAA----GGAGAC--------AGCACA---GCAUAAGUAAAUGUUGGCAAAUAAAUAUGCAUGGCCGAAUGGAGAGAUGGCCAA-AAGAUGGUGUUC ...........((((((((((((...----(....)--------.((.((---((((......)))))))).............))))).........)))))))..-............ ( -29.00) >DroSim_CAF1 91103 105 - 1 UUGAGUUCGACGGCCAUUGGCCACAA----GGAGAC--------AGCACA---GCAUAAGUAAAUGUUGGCAAAUAAAUAUGCAUGGCCGAAUGGCGAGAUGGCCAAAAAGAUGGUGUUC ........(((.(((((((((((...----(....)--------.((.((---((((......)))))))).............))))).))))))......((((......))))))). ( -32.50) >DroEre_CAF1 101261 104 - 1 UUGAGUU----GGCCAU-GGCCAAGA----AGCGGCUGGCCGCAAAUGCA---GCAUAAGUAAAUGUUGGCAAAUAAAUAUGCAUGGCCGAAUGGAG--AUACCCGAACA--UGGCACAC ......(----((((((-(((((...----.((((....)))).....((---((((......))))))(((........))).)))))...(((..--....)))...)--)))).)). ( -33.10) >DroYak_CAF1 103461 107 - 1 UUGAGUU----GGCCAU-GGCCGAGACCAGAACGACUUGCCUCAAAUGCA---GCAUAAGUAAAUGUUGGCAAAUAAAUAUGCAUGGCCGAAUGGAG--AUACCCGAACA--CGGCACA- .....((----((((((-(...(((.(.((.....)).).)))...((((---((((......))))).)))..........)))))))))......--....(((....--)))....- ( -25.70) >consensus UUGAGUUCGACGGCCAUUGGCCAAAA____GGAGAC________AGCACA___GCAUAAGUAAAUGUUGGCAAAUAAAUAUGCAUGGCCGAAUGGAGAGAUGGCCAAACA_AUGGUGCUC ............(((.(((((((........................(((...((....))...)))..(((........))).))))))).(((........))).......))).... (-19.20 = -18.72 + -0.48)

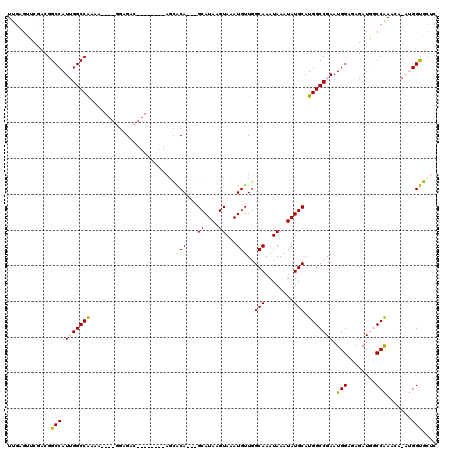
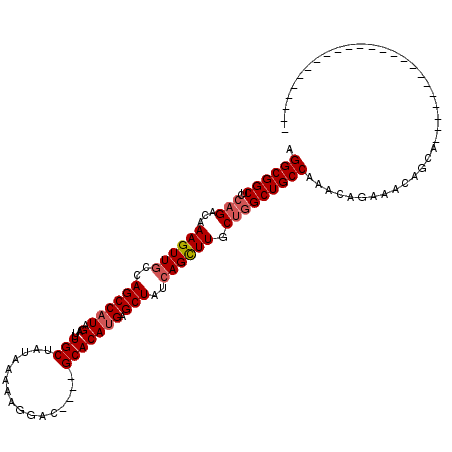
| Location | 17,214,286 – 17,214,378 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17214286 92 + 23771897 UGGCGGCUCCAGACAAAGUUACCAGCCAUAGAUUUGCUCUAAAAAGGAC----GCACAUGAGCUAUCAGCUUGCUGGCUGCCAAACCGAAACAGCA------------------------ (((((((..(((...(((((...((((((.(...((((((.....))).----))))))).)))...))))).)))))))))).............------------------------ ( -24.90) >DroSec_CAF1 97999 92 + 1 AGGCGGCUCCAGACAAAGUUGCCAGCCAUAGAUUUGCUUUAAAAAGGAC----GCACAUGAGCUAUCAGCUUGCUGGCUGCCAAACAGAAACAGCA------------------------ .((((((..(((...((((((..((((((.(...(((.((......)).----))))))).)))..)))))).)))))))))..............------------------------ ( -25.70) >DroSim_CAF1 91208 92 + 1 AGGCGGCUCCAGACAAAGUUGCCAGCCAUAGAUUUGCUUUAAAAAGGAC----GCACAUGAGCUAUCAGCUUGCUGGCUGCCAAACAGAAACAGCA------------------------ .((((((..(((...((((((..((((((.(...(((.((......)).----))))))).)))..)))))).)))))))))..............------------------------ ( -25.70) >DroEre_CAF1 101365 89 + 1 AGGCGGCUCCAGACAAAGUUGCCAGCCAUGGAUUUGCCAUA---AGGACAUAAGCACAUGAGCUAUCAGUUUGCCUGCUGCCAAACAGCAAU---------------------------- .((((((..(((((.....((..((((((((...((((...---.)).))....).)))).)))..)))))))...))))))..........---------------------------- ( -24.90) >DroYak_CAF1 103568 113 + 1 AGGCGGCUCCAGACAAAGUUGCCAGCCAUGGAUUUGCCAUA---AGGAC----GCACACGAGCUAUCAGUUUGCUGGCUGCCAAACACCAACACCAACAGCAAUAGCAACCAACAGACUU .(((((((.(((((...(((((...((((((.....)))).---.))..----))).))((....)).)))))..))))))).................((....))............. ( -26.90) >consensus AGGCGGCUCCAGACAAAGUUGCCAGCCAUAGAUUUGCUAUAAAAAGGAC____GCACAUGAGCUAUCAGCUUGCUGGCUGCCAAACAGAAACAGCA________________________ .((((((..(((...((((((..((((((.(...(((................))))))).)))..)))))).)))))))))...................................... (-19.19 = -19.75 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:15 2006