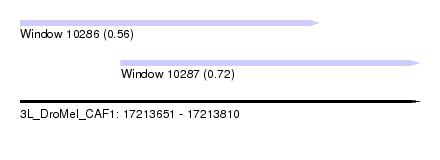
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,213,651 – 17,213,810 |
| Length | 159 |
| Max. P | 0.716046 |

| Location | 17,213,651 – 17,213,770 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -20.42 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213651 119 + 23771897 UAGUUGCUGUAAAAUGCUGCUGCUCUUAAUCACCAAACAGAAUGGUGUAGUU-UUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUUCCUUAUAAAUCAAACUGCCUCCAGGGACACAAA ((((.((........)).))))........(((((.......)))))..(((-((((...(((((((((((...((.((.......)).)).))))))))))).....)))))))..... ( -23.60) >DroSec_CAF1 97362 118 + 1 UUGUUGCUGUAAAAUG--GCUGCUCUUAAUCACCGAACAGAAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUCCGUAAUAAAUCAAACUGCCUACAGGGACACAAA .((((.((((((((..--(((((.((...((........))..)).)))))..)))....(((((((((((....(((.........)))..)))))))))))...))))).)))).... ( -28.30) >DroSim_CAF1 89840 118 + 1 UAGUUGCUGUAAAAUG--GCUGCUCUUAAUCACCGAACAGAAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUCCGUAAUAAAUCAAACUGCCUACAGGGACACAAA ..(((.((((((((..--(((((.((...((........))..)).)))))..)))....(((((((((((....(((.........)))..)))))))))))...))))).)))..... ( -27.90) >DroEre_CAF1 100740 118 + 1 UAGUUGCUAAGACAUG--GCUGCUCUUAAUCACCAAACAGAAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUCAUUUCUUAAUAAAUCACACUGCCUCCAGGGGCACAAA ..........((((.(--(((((.((...((........))..)).))))))...)))).(((.((((((((((((....))))).......))))))).)))((((....))))..... ( -26.11) >DroYak_CAF1 102940 118 + 1 UAGUUGCUGUAAAAUG--UCUGCUCUUAAUCACCAAACGGAGUGGUGUAGUUUUUUCGCAAGUUUGAUUUAAUGAACGACUUAAUUUCUUAAUAAAUCAAACUACUUGCAGGGACAUAAA ..(((.((((((....--..(((.......(((((.......)))))..........)))(((((((((((.(((.............))).)))))))))))..)))))).)))..... ( -26.55) >consensus UAGUUGCUGUAAAAUG__GCUGCUCUUAAUCACCAAACAGAAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUUCGUAAUAAAUCAAACUGCCUACAGGGACACAAA ..(((.(((((.......(((((.((...((........))..)).))))).........(((((((((((.....................)))))))))))...))))).)))..... (-20.42 = -21.02 + 0.60)

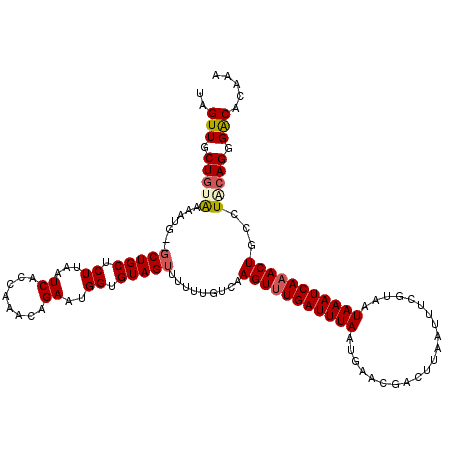
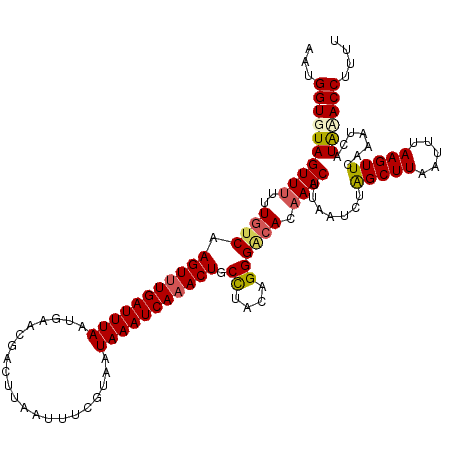
| Location | 17,213,691 – 17,213,810 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213691 119 + 23771897 AAUGGUGUAGUU-UUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUUCCUUAUAAAUCAAACUGCCUCCAGGGACACAAACUUAAUUUAGCUUAAUUUAAGUUCAAAUCAUAUACCUUUU ...(((((((((-(.((((.(((((((((((...((.((.......)).)).))))))))))).((....)))))).))))...((((((((((...)))))).))))..)))))).... ( -25.10) >DroSec_CAF1 97400 120 + 1 AAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUCCGUAAUAAAUCAAACUGCCUACAGGGACACAAACUUAAUCUAGCUUAAUUUAAGUUCAAAUCAUAAACCUUUU ...(((..(((((..((((.(((((((((((....(((.........)))..))))))))))).((....)))))).)))))......((((((...))))))..........))).... ( -21.10) >DroSim_CAF1 89878 120 + 1 AAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUCCGUAAUAAAUCAAACUGCCUACAGGGACACAAACUUAAUCUAGCUUAAUUUAAGUUCAAAUCAUAAACCUUUU ...(((..(((((..((((.(((((((((((....(((.........)))..))))))))))).((....)))))).)))))......((((((...))))))..........))).... ( -21.10) >DroEre_CAF1 100778 120 + 1 AAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUCAUUUCUUAAUAAAUCACACUGCCUCCAGGGGCACAAACUUAAUCUAGCUUGAUUAAAGUUCACCUUAUGCACCUGCU ...((((((....(((((..(((.((((((((((((....))))).......))))))).)))((((....))))))))).((((((......))))))...........)))))).... ( -27.41) >DroYak_CAF1 102978 120 + 1 AGUGGUGUAGUUUUUUCGCAAGUUUGAUUUAAUGAACGACUUAAUUUCUUAAUAAAUCAAACUACUUGCAGGGACAUAAACCUAAUCUGGCUUGAUUAAAGUUCACCUCAUGUACCUCUU .(.((((.(.(((..((((((((((((((((.(((.............))).))))))))...))))))(((........)))..........))..))).).)))).)........... ( -24.72) >consensus AAUGGUGUAGUUUUUUGUCAAGUUUGAUUUAAUGAACGACUUAAUUUCGUAAUAAAUCAAACUGCCUACAGGGACACAAACUUAAUCUAGCUUAAUUUAAGUUCAAAUCAUAAACCUUUU ...((((((((((..((((.(((((((((((.....................))))))))))).((....)))))).)))).......(((((.....))))).......)))))).... (-17.98 = -18.42 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:05 2006