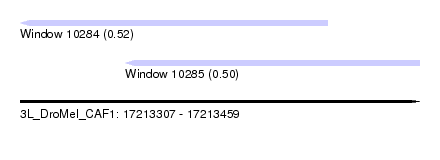
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,213,307 – 17,213,459 |
| Length | 152 |
| Max. P | 0.519672 |

| Location | 17,213,307 – 17,213,424 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -32.04 |
| Energy contribution | -32.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213307 117 - 23771897 GUUAGUCAAAUGACAGAG---GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUCCUGCCGCAGGCCACUCAGCCAACUGCAGAUGUAAGACCAAUGUGUCGUAUGCGCAACGUGGA ....(((....)))....---..((.((..(((((((((((...((.....(((((..((((.(..(((......)))..).))))..)))))..))...)).)))))))))..)).)). ( -36.70) >DroSec_CAF1 97011 120 - 1 GGUAGUCAAACGACAGAGGAGAGCCGCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAACUGCAGAUGUGAGUCCAAUGUGUCGUAUGCGCAACGUGGA ....(((....))).........(((((..(((((((((((....((..((((((((((....)))(((......)))....))))).))...)).....)).)))))))))..))))). ( -40.30) >DroSim_CAF1 89489 120 - 1 GGUAGUCAAACGACAGAGGAGGGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAACUGCAGAUGUGAGUCCAAUGUGUCGUAUGCGCAACGUGGA (((.(((....))).(((...(((((.(.((((((...(((((.(....).)))))))).))).)))))).))).)))...............((((.(((((.....)))))...)))) ( -39.60) >DroEre_CAF1 100384 117 - 1 GGUAGUCAAAUGACAGAG---GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAACUGCAGAUCCGAGACCAAUGUGUCGUAUGCGCAACGUAGA (((.(((....))).(((---(((((.(.((((((...(((((.(....).)))))))).))).)))))).))).)))....(((....(((.((....)).)))..))).......... ( -39.70) >DroYak_CAF1 102563 117 - 1 GGUAGUCAAAUGACAGAG---GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUCCUGCUGCCGCAGGCCACUCAGCCAACUGCAGAUUCGAGACCAAUGUGUCGUAUGCGCAACGUAGA (((.(((....))).(((---(((((((.(((((....)).....(((....)))..)))..)).))))).))).)))....(((....(((.((....)).)))..))).......... ( -34.60) >consensus GGUAGUCAAAUGACAGAG___GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAACUGCAGAUGUGAGACCAAUGUGUCGUAUGCGCAACGUGGA ....(((....))).........((.((..(((((((((((...((....(((.....((((.(..(((......)))..).))))....)))..))...)).)))))))))..)).)). (-32.04 = -32.28 + 0.24)


| Location | 17,213,347 – 17,213,459 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -26.42 |
| Energy contribution | -27.62 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17213347 112 - 23771897 AACGUGGAUUAAGACCC-----CCCCCGUUCCCAGUUUUGGUUAGUCAAAUGACAGAG---GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUCCUGCCGCAGGCCACUCAGCCAA ((((.((..........-----.)).)))).......((((((.(((....))).(((---(((((.(.((((.....(((((.(....).)))))...)))).)))))).))))))))) ( -34.60) >DroSec_CAF1 97051 120 - 1 AACGUGGAUUAAGACCCCACCCCCCUCGUUCCCAGUUUUGGGUAGUCAAACGACAGAGGAGAGCCGCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAA ...((((................((((...(((......)))..(((....))).))))...(((....)))(((...(((((.(....).))))))))..)))).(((......))).. ( -36.20) >DroSim_CAF1 89529 120 - 1 AACGUGGAUUAAGACCCCAACCCCCUCGUUCCCAGUUUUGGGUAGUCAAACGACAGAGGAGGGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAA ...((((.............(((((((...(((......)))..(((....))).)))).)))(((.(.((((((...(((((.(....).)))))))).))).))))))))........ ( -38.40) >DroEre_CAF1 100424 108 - 1 AACGUGGAUUAAAACCC--------CCGAUCCCAGUU-UGGGUAGUCAAAUGACAGAG---GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAA ....(((.........(--------((((.......)-))))..(((....))).(((---(((((.(.((((((...(((((.(....).)))))))).))).)))))).)))..))). ( -36.70) >DroYak_CAF1 102603 108 - 1 AACGUGGAUUAAAACC---------CCGAUCCGAGUUUUGGGUAGUCAAAUGACAGAG---GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUCCUGCUGCCGCAGGCCACUCAGCCAA (((.((((((......---------..)))))).)))...(((.(((....))).(((---(((((((.(((((....)).....(((....)))..)))..)).))))).))).))).. ( -32.50) >consensus AACGUGGAUUAAGACCC_____CCCCCGUUCCCAGUUUUGGGUAGUCAAAUGACAGAG___GGCCUCGAGGCGCAUAUGCAAAAUGAAAACUUUGCUGCUGCCGCAGGCCACUCAGCCAA ....(((.......................(((......)))..(((....))).(((...(((((...((((((...(((((.(....).)))))))).)))..))))).)))..))). (-26.42 = -27.62 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:03 2006