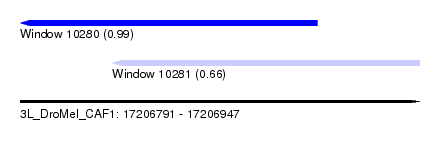
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,206,791 – 17,206,947 |
| Length | 156 |
| Max. P | 0.993709 |

| Location | 17,206,791 – 17,206,907 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -43.67 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.03 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
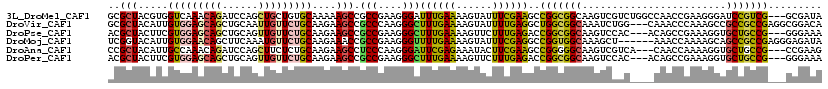
>3L_DroMel_CAF1 17206791 116 - 23771897 GCGCUACGUGGUCAAACAGAUCCAGCUGCUGUGCAAAAAGCCGCCGAAGGGAUUUGAAAAGUAUUUCGAAGCCGGCGGCAAGUCGUCUGGCCAACCGAAGGGAUCCGUCG---GCGAUA .(((((((((((((.((.(((...(((((((.((.........((....)).(((((((....))))))))))))))))..))))).)))))).((....))...))).)---)))... ( -42.80) >DroVir_CAF1 113128 116 - 1 GCGCUACAUUGUGGAGCAGCUGCAAUUGUUCUGCAAGAAGCCGCCCAAGGGCUUUGAAAAGUAUUUUGAGGCUGGCGGCAAAUCUGG---CAAACCCAAAGCCGCCGCCGAGGCGGACA ..((((..(((..((((((......))))))..)))...((((((....(((((..(((....)))..))))))))))).....)))---).........((((((.....))))).). ( -46.60) >DroPse_CAF1 96928 113 - 1 ACGCUACUUCGUGGAGCAGCUGCAGUUGUUCUGCAAGAAGCCGCCGAAGGGCUUUGAAAAGUUCUUUGAGACCGGCGGCAAGUCCAC---ACAGCCGAAAGGUGCUGCCG---GGGAAA ..(((.(((.(..(((((((....)))))))..)))).)))...((((((((((....))))))))))...((((((((..((....---)).(((....))))))))))---)..... ( -50.00) >DroMoj_CAF1 111800 113 - 1 UCGGUACAUUGUGGAACAGCUUCAAAUGUUCUGCAAGAAACCGCCGAAGGGUUUUGAAAAGUAUUUCGAGGCCGGUGGCAAAGCU------AAACCAAAAGCAGCCGCCGAGGGAGAUA .((((...(((..(((((........)))))..)))...))))((....((((((((((....))))))))))((((((...(((------........))).))))))..))...... ( -42.50) >DroAna_CAF1 73166 113 - 1 CCGCUACAUUGCCAAACAGAUCCAGCUUCUCUGCAAGAAGCCUCCCAAGGGAUUCGAGAAAUACUUCGAAGCCGGGGGCAAGUCGUCA---CAACCAAAAGGUGCUGCCG---CCGAAG ..((.((.(((((...........((((((.....))))))..(((...((.((((((......)))))).)))))))))))).))..---.........((((....))---)).... ( -30.10) >DroPer_CAF1 69688 113 - 1 ACGCUACUUCGUGGAGCAGCUGCAGUUGUUCUGCAAGAAGCCGCCGAAGGGCUUUGAAAAGUUCUUUGAGACCGGCGGCAAGUCCAC---ACAGCCGAAAGGUGCUGCCG---GGGAAA ..(((.(((.(..(((((((....)))))))..)))).)))...((((((((((....))))))))))...((((((((..((....---)).(((....))))))))))---)..... ( -50.00) >consensus ACGCUACAUUGUGGAACAGCUGCAGUUGUUCUGCAAGAAGCCGCCGAAGGGCUUUGAAAAGUACUUCGAGGCCGGCGGCAAGUCCAC____CAACCGAAAGGUGCCGCCG___GGGAAA ..(((.....(((((((((......)))))))))....))).(((....)))((((((......))))))..(((((((........................)))))))......... (-23.10 = -23.03 + -0.08)
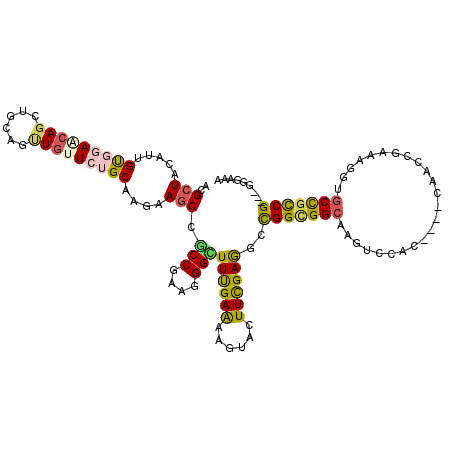
| Location | 17,206,827 – 17,206,947 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -44.91 |
| Consensus MFE | -28.55 |
| Energy contribution | -26.61 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17206827 120 - 23771897 CCGGGGUUUACAUCCUGCCCACGGAGCUGUCGCCCCUGCUGCGCUACGUGGUCAAACAGAUCCAGCUGCUGUGCAAAAAGCCGCCGAAGGGAUUUGAAAAGUAUUUCGAAGCCGGCGGCA ...((((((......((.(((((.(((.((.((....)).))))).))))).))...)))))).(((((((.((.........((....)).(((((((....)))))))))))))))). ( -41.30) >DroPse_CAF1 96961 120 - 1 UCGGCGUUGGUAUGCUGCCGGGGCAACUUACGCCCCUAAUACGCUACUUCGUGGAGCAGCUGCAGUUGUUCUGCAAGAAGCCGCCGAAGGGCUUUGAAAAGUUCUUUGAGACCGGCGGCA ..(((((.(((.....)))(((((.......)))))....))))).(((.(..(((((((....)))))))..))))..(((((((((((((((....))))))))......))))))). ( -53.10) >DroSec_CAF1 91309 120 - 1 CCGGUGUUUACAUCAUGCCCACGGAGCUGUCGCCUCUGCUGCGCUACGUGGUCAAACAGAUCCAGCUGCUGUGCAAGAAGCCGCCGAAGGGAUUCGAAAAGUAUUUCGAAGCCGGCGGCA ..((((....)))).((.(((((.(((.((.((....)).))))).))))).))..........(((((((.((.........((....)).(((((((....)))))))))))))))). ( -42.80) >DroSim_CAF1 83593 120 - 1 CCGGGGUUUACAUCAUGCCCACGGAGCUGUCGCCUCUGCUGCGCUACGUGGUCAAACAGAUCCAGCUGCUGUGCAAGAAGCCGCCGAAGGGAUUCGAAAAGUAUUUCGAAGCCGGCGGCA ...((((((......((.(((((.(((.((.((....)).))))).))))).))...)))))).(((((((.((.........((....)).(((((((....)))))))))))))))). ( -43.40) >DroAna_CAF1 73199 120 - 1 CUGGAGUCUACAUGUUCCCAGUGGAGCUGAAUCCGCUUAUCCGCUACAUUGCCAAACAGAUCCAGCUUCUCUGCAAGAAGCCUCCCAAGGGAUUCGAGAAAUACUUCGAAGCCGGGGGCA ..((((.(.....)))))(((.(((((((.(((.........((......))......))).))))))).)))......((((((....((.((((((......)))))).)))))))). ( -35.76) >DroPer_CAF1 69721 120 - 1 UCGGCGUUGGUAUGCUGCCGGGGCAACUUACGCCCCUAAUACGCUACUUCGUGGAGCAGCUGCAGUUGUUCUGCAAGAAGCCGCCGAAGGGCUUUGAAAAGUUCUUUGAGACCGGCGGCA ..(((((.(((.....)))(((((.......)))))....))))).(((.(..(((((((....)))))))..))))..(((((((((((((((....))))))))......))))))). ( -53.10) >consensus CCGGCGUUUACAUCCUGCCCACGGAGCUGACGCCCCUGAUGCGCUACGUGGUCAAACAGAUCCAGCUGCUCUGCAAGAAGCCGCCGAAGGGAUUCGAAAAGUACUUCGAAGCCGGCGGCA .((((((....)))))).(((((.(((...............))).))))).............((......)).....((((((....((.((((((......)))))).)))))))). (-28.55 = -26.61 + -1.94)

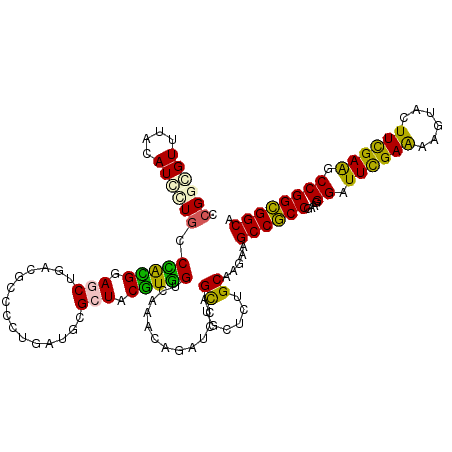
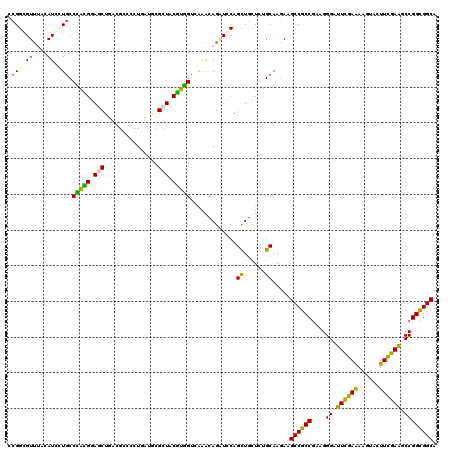
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:59 2006