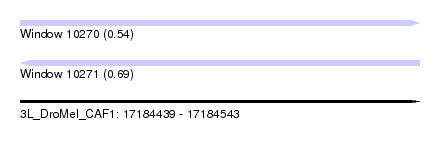
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,184,439 – 17,184,543 |
| Length | 104 |
| Max. P | 0.689949 |

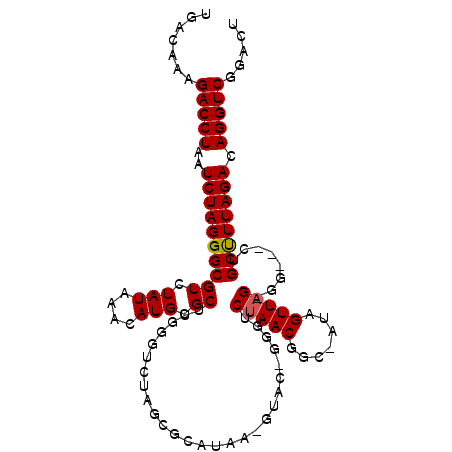
| Location | 17,184,439 – 17,184,543 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.87 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17184439 104 + 23771897 AGUCCGACCUGUCUAAAGCAG---CC-CUAACUAU-GCCGUUAGACCCUGUACGUUAUGCGCUAGAGCCGAGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA .....((((((((((..((..---..-(((((...-...)))))...(((((...((((((((.......))))))))..)))))..))..)))).))))))....... ( -33.20) >DroVir_CAF1 82765 92 + 1 --GCCGACCUGUCUAAGGCAGGCACC-CAAACUAUUGCAGUUAGACU--------------UUAGAAACCGGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA --...((((((((((.(((..((.((-....(((..(........).--------------.))).....)).))..(((....)))))).)))).))))))....... ( -23.80) >DroPse_CAF1 72281 99 + 1 UCGUCGACCUGUCUAAAGCAG---CCUCUAACUAU-GCUGUUAGACCCCGCCC-CCACGCC-----CCCAUGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA .....((((((((((..((.(---(.((((((...-...))))))........-....((.-----.....))))..(((....)))))..)))).))))))....... ( -23.30) >DroEre_CAF1 71004 104 + 1 AGUCCGACCUGUCUAAAGCAG---CC-CUAACUAU-GCCGUUAGACCCUGUACGUUAUGCGCUAGACCCAAGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA .....((((((((((..((..---..-(((((...-...)))))...(((((...((((((((.......))))))))..)))))..))..)))).))))))....... ( -33.10) >DroYak_CAF1 72370 104 + 1 AGUCGGACCUGUCUAAAGCAG---CC-CUAACUAU-GCCGUUAGACCCAGUACGUUAUGCGCUAGACCCAAGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA ....(((((((((((..((..---..-(((((...-...))))).....((....((((((((.......)))))))).......))))..)))).)))))))...... ( -29.80) >DroAna_CAF1 50733 90 + 1 AGUCCGACCUGUCUAAAGCAG---CC-CUAACUAU-GCCGUUAGAC--------------GUUAGAACCGAGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA .....((((((((((..((.(---(.-(((((...-...)))))..--------------(((.......)))))..(((....)))))..)))).))))))....... ( -20.60) >consensus AGUCCGACCUGUCUAAAGCAG___CC_CUAACUAU_GCCGUUAGACCC_GUAC_UUAUGCGCUAGACCCAAGCGCAUGUUUAUAGACGCCCUAGAUUAGGUCUUUGUCA .....((((((((((..((........(((((.......)))))..............(((((.......)))))............))..)))).))))))....... (-18.64 = -19.87 + 1.22)

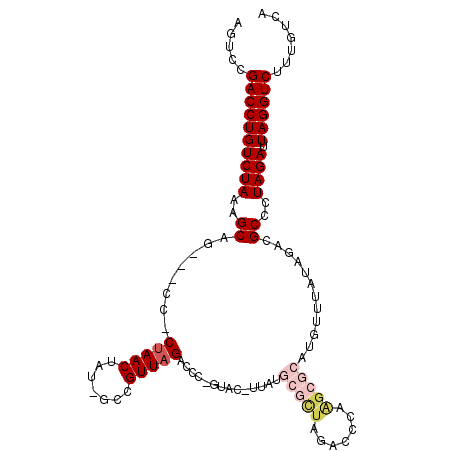
| Location | 17,184,439 – 17,184,543 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17184439 104 - 23771897 UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCUCGGCUCUAGCGCAUAACGUACAGGGUCUAACGGC-AUAGUUAG-GG---CUGCUUUAGACAGGUCGGACU .......(((((..((((((((..........(((((((.......)))))))........((.((((((...-...)))))-).---)))))))))).)))))..... ( -35.10) >DroVir_CAF1 82765 92 - 1 UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCCGGUUUCUAA--------------AGUCUAACUGCAAUAGUUUG-GGUGCCUGCCUUAGACAGGUCGGC-- .......(((((..((((((((((......))..(((((.(...(((.--------------.((......))..)))..).-)))))..)))))))).)))))...-- ( -28.70) >DroPse_CAF1 72281 99 - 1 UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCAUGGG-----GGCGUGG-GGGCGGGGUCUAACAGC-AUAGUUAGAGG---CUGCUUUAGACAGGUCGACGA .......(((((..((((((((((((.....(((((.(....)-----.))))).-))))((..((((((...-...))))))..---)))))))))).)))))..... ( -35.50) >DroEre_CAF1 71004 104 - 1 UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCUUGGGUCUAGCGCAUAACGUACAGGGUCUAACGGC-AUAGUUAG-GG---CUGCUUUAGACAGGUCGGACU .......(((((..((((((((..........(((((((.......)))))))........((.((((((...-...)))))-).---)))))))))).)))))..... ( -35.90) >DroYak_CAF1 72370 104 - 1 UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCUUGGGUCUAGCGCAUAACGUACUGGGUCUAACGGC-AUAGUUAG-GG---CUGCUUUAGACAGGUCCGACU .......(((((..((((((((((((.(((..((((..((((..((((..(.....)..))))..))))..))-))..))))-))---).)))))))).)))))..... ( -37.40) >DroAna_CAF1 50733 90 - 1 UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCUCGGUUCUAAC--------------GUCUAACGGC-AUAGUUAG-GG---CUGCUUUAGACAGGUCGGACU .......(((((..((((((((((..........))))))(((((((((--------------(((....)))-...)))))-))---))....)))).)))))..... ( -29.30) >consensus UGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCUUGGGUCUAGCGCAUAA_GUAC_GGGUCUAACGGC_AUAGUUAG_GG___CUGCUUUAGACAGGUCGGACU .......(((((..((((((((((.(((....))).))...........................(((((.......)))))........)))))))).)))))..... (-19.32 = -19.35 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:49 2006