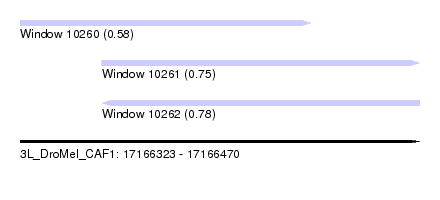
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,166,323 – 17,166,470 |
| Length | 147 |
| Max. P | 0.784064 |

| Location | 17,166,323 – 17,166,430 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.98 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -8.60 |
| Energy contribution | -10.28 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17166323 107 + 23771897 AAAUCUGUUUACUGAC-------AAAAU---UCUUGAUCUCUAGCUGAUUUCAUUUACACAAUAGACAUUGAUGUCGAUAUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCA ......((((((((.(-------((((.---((.(((..((.....))..)))...........((((....))))))...)))))))))))))(((((((....)))))))..... ( -26.50) >DroSec_CAF1 50732 92 + 1 AAAUCUGUUCACUGAC-------AAAAUAGAUA--GAUCUCUAGCUGAUUUCAUUUACACAAU----------------AUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCA ......((((((((.(-------((((((((((--((((.......))))).)))))......----------------..)))))))))))))(((((((....)))))))..... ( -24.20) >DroSim_CAF1 42750 110 + 1 AAAUCUGUUUACUGAC-------AAAAUAGAUCCUAAUCUCUAGCUGAUUUCAUUUACACAAUAGACAAUGAUGUCGACAUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCA ......((((((((.(-------(((((((((...((((.......))))..))))).......((((....)))).....)))))))))))))(((((((....)))))))..... ( -25.50) >DroEre_CAF1 52900 92 + 1 ACAUCUGUGAACUUAC-------UAAGU---AGUUGAAUUCUAG--------------AUAAUAGACAUUGAUAUCGAUUU-UUUCCAGUGAGCCUGCAGUUACCAUUGCAGUUGCA ......(..(.(((((-------(....---.((..(..((((.--------------....))))..)..))...((...-..)).)))))).(((((((....))))))))..). ( -16.10) >DroYak_CAF1 53907 104 + 1 AGAACUCCUACCUUACAAAUAAAUAAAU---ACCCGAACUCUAG----------UUACAUAAGAGAUAUUGAUAUCGAUUUUUUUCCAGUGCACCUGCAGUUACCAUUGCAGUUGCA ............................---.............----------........((((.((((....))))...))))...((((.(((((((....))))))).)))) ( -15.30) >consensus AAAUCUGUUAACUGAC_______AAAAU___UCCUGAUCUCUAGCUGAUUUCAUUUACACAAUAGACAUUGAUAUCGAUAUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCA ......((((((((.....................(....).......................((((....))))..........))))))))(((((((....)))))))..... ( -8.60 = -10.28 + 1.68)

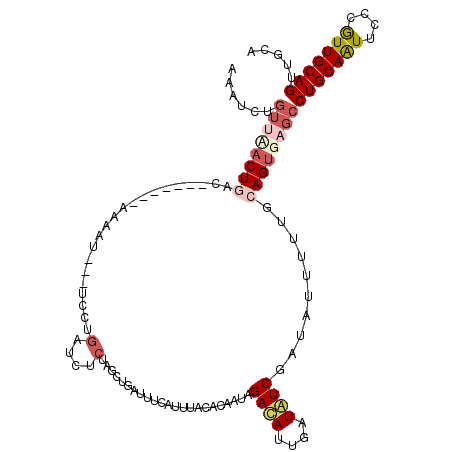

| Location | 17,166,353 – 17,166,470 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17166353 117 + 23771897 CUAGCUGAUUUCAUUUACACAAUAGACAUUGAUGUCGAUAUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCACUUGCAUUUGCAAUUUGCAUUUCGGACGCUCAGGCAAUUA ........................((((....)))).......((((..((((((((((((....))))))).((((.((((....))))..))))........))))).))))... ( -29.10) >DroSec_CAF1 50763 101 + 1 CUAGCUGAUUUCAUUUACACAAU----------------AUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCACUUGCAUUUGCAAUUUGCAUUUCGGACGCUCAGGCAAUUA .......................----------------....((((..((((((((((((....))))))).((((.((((....))))..))))........))))).))))... ( -25.80) >DroSim_CAF1 42783 117 + 1 CUAGCUGAUUUCAUUUACACAAUAGACAAUGAUGUCGACAUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCACUUGCAUUUGCAAUUUGCAUUUCGGACGCUCAGGCAAUUA ...(.((((.(((((............))))).)))).)....((((..((((((((((((....))))))).((((.((((....))))..))))........))))).))))... ( -30.00) >DroEre_CAF1 52930 102 + 1 CUAG--------------AUAAUAGACAUUGAUAUCGAUUU-UUUCCAGUGAGCCUGCAGUUACCAUUGCAGUUGCACUUGCAUUUGCAAUUUGCAUUUCGGACGCUCAGGCAAUUA ....--------------........(((((..........-....))))).(((((.(((..((..((((((((((........))))).)))))....))..))))))))..... ( -24.94) >DroYak_CAF1 53944 107 + 1 CUAG----------UUACAUAAGAGAUAUUGAUAUCGAUUUUUUUCCAGUGCACCUGCAGUUACCAUUGCAGUUGCACUUGCAUUUGCAAUUUGCAUUUCGGACGCUCAGGCAAUUA ....----------........((((.((((....)))).))))(((((((((.(((((((....))))))).))))))((((.........))))....))).((....))..... ( -26.30) >consensus CUAGCUGAUUUCAUUUACACAAUAGACAUUGAUAUCGAUAUUUUUGCAGUGAGCCUGCAAUUCCCGUUGCAGUUGCACUUGCAUUUGCAAUUUGCAUUUCGGACGCUCAGGCAAUUA ...........................................((((..((((((((((((....))))))).((((.((((....))))..))))........))))).))))... (-21.58 = -21.90 + 0.32)


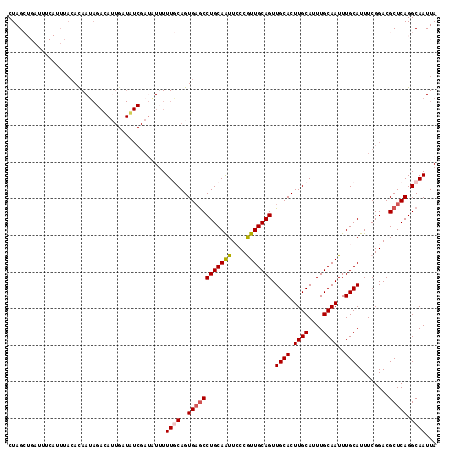
| Location | 17,166,353 – 17,166,470 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17166353 117 - 23771897 UAAUUGCCUGAGCGUCCGAAAUGCAAAUUGCAAAUGCAAGUGCAACUGCAACGGGAAUUGCAGGCUCACUGCAAAAAUAUCGACAUCAAUGUCUAUUGUGUAAAUGAAAUCAGCUAG ...((((.(((((........((((..((((....)))).)))).((((((......)))))))))))..)))).......((((....))))........................ ( -28.70) >DroSec_CAF1 50763 101 - 1 UAAUUGCCUGAGCGUCCGAAAUGCAAAUUGCAAAUGCAAGUGCAACUGCAACGGGAAUUGCAGGCUCACUGCAAAAAU----------------AUUGUGUAAAUGAAAUCAGCUAG ...((((.(((((........((((..((((....)))).)))).((((((......)))))))))))..))))....----------------....................... ( -25.80) >DroSim_CAF1 42783 117 - 1 UAAUUGCCUGAGCGUCCGAAAUGCAAAUUGCAAAUGCAAGUGCAACUGCAACGGGAAUUGCAGGCUCACUGCAAAAAUGUCGACAUCAUUGUCUAUUGUGUAAAUGAAAUCAGCUAG ...((((.(((((........((((..((((....)))).)))).((((((......)))))))))))..)))).......((((....))))........................ ( -29.50) >DroEre_CAF1 52930 102 - 1 UAAUUGCCUGAGCGUCCGAAAUGCAAAUUGCAAAUGCAAGUGCAACUGCAAUGGUAACUGCAGGCUCACUGGAAA-AAAUCGAUAUCAAUGUCUAUUAU--------------CUAG ........(((((........((((..((((....)))).)))).(((((........))))))))))(((((..-.(((.((((....)))).))).)--------------)))) ( -25.60) >DroYak_CAF1 53944 107 - 1 UAAUUGCCUGAGCGUCCGAAAUGCAAAUUGCAAAUGCAAGUGCAACUGCAAUGGUAACUGCAGGUGCACUGGAAAAAAAUCGAUAUCAAUAUCUCUUAUGUAA----------CUAG ...((((.((((..(((....(((.....)))......((((((.(((((........))))).)))))))))........((((....)))).)))).))))----------.... ( -26.40) >consensus UAAUUGCCUGAGCGUCCGAAAUGCAAAUUGCAAAUGCAAGUGCAACUGCAACGGGAAUUGCAGGCUCACUGCAAAAAUAUCGACAUCAAUGUCUAUUGUGUAAAUGAAAUCAGCUAG ...((((.(((((........((((..((((....)))).)))).((((((......)))))))))))..)))).......((((....))))........................ (-21.86 = -22.62 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:40 2006