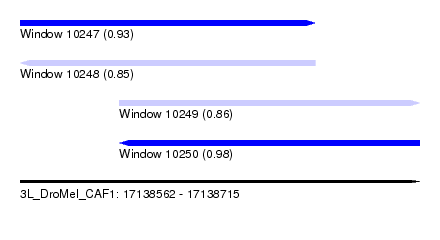
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,138,562 – 17,138,715 |
| Length | 153 |
| Max. P | 0.980546 |

| Location | 17,138,562 – 17,138,675 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -22.72 |
| Energy contribution | -21.96 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17138562 113 + 23771897 ACAAAAAUGCAUCCUUGAACACGCACACUUAACUGGGCCUGCGUGAGUGGCAACCUCUUGCCACCCACAC-UACCCAACCCCCUCGGACGCCAUGGA-ACCACCGCCUCUCCACU ........((.(((.....((((((..(((....)))..)))))).(((((((....)))))))......-..............))).))..((((-...........)))).. ( -27.00) >DroSec_CAF1 23002 113 + 1 ACAAAAAUGCAUCCUUGAACACGCACACUUAACUGGGCCUGCGUGAGUGGCAACCUCUUGCCACCCACAC-UACCCACCCCCCUUGGACGCCUUGGA-CCCACCGCCUCUUCACU .......(((............)))........((((((.(((((.(((((((....))))))).)))..-...(((.......)))..))...)).-))))............. ( -26.80) >DroEre_CAF1 24064 113 + 1 ACAAAAAUGCAUCCUUGAACACGCACACUUAACUGGGCCUGCGUGAGUGGCAACCUCUUGCCACCCACGA-AACCCACCCCCUUUGGACUCCUUGGA-CCCAACGCCUCUUCACU ...............((((...((.........((((....((((.(((((((....))))))).)))).-..))))......((((..((....))-.)))).))...)))).. ( -29.10) >DroYak_CAF1 25253 114 + 1 ACAAAAAUGCAUCCUUGAACACGCACACUUAACUGGGCCUGCGUGAGUGGCAACCUCUUGCCACCCGCAGUUCCCCACCCCCUUUGGACUCCUUGGA-CCCACCGCCUCUCCACU .......(((............)))........((((.(((((.(.(((((((....)))))))))))))...)))).......((((.....(((.-....)))....)))).. ( -28.20) >DroAna_CAF1 5217 110 + 1 ACAAAAAUGCAUCCUUGAACACGCACACUUAACUGGGCCUGCGUGAGUGGCAGCCUCUUGCCUCCCUCGA-C----AGCCCGCCAGGAUCCUUUCUUCAAGACUGUCCCACCGAU ........(((..(((((................((((.((((.(.(.(((((....))))).).).)).-)----)))))...((((....)))))))))..)))......... ( -25.00) >consensus ACAAAAAUGCAUCCUUGAACACGCACACUUAACUGGGCCUGCGUGAGUGGCAACCUCUUGCCACCCACAA_UACCCACCCCCCUUGGACGCCUUGGA_CCCACCGCCUCUCCACU ........((.(((.....((((((..(((....)))..)))))).(((((((....))))))).....................))).....(((...)))..))......... (-22.72 = -21.96 + -0.76)

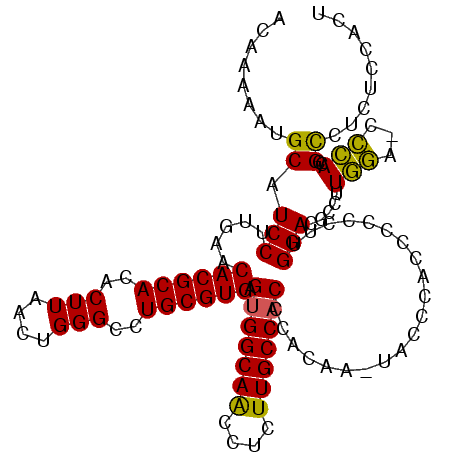

| Location | 17,138,562 – 17,138,675 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -29.36 |
| Energy contribution | -28.80 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17138562 113 - 23771897 AGUGGAGAGGCGGUGGU-UCCAUGGCGUCCGAGGGGGUUGGGUA-GUGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCCCAGUUAAGUGUGCGUGUUCAAGGAUGCAUUUUUGU ....(.((.((.(((..-..))).)).))).......((((((.-(((((((((((((....)))))))))))))..)))))).((((.((((((.(....).)))))).)))). ( -46.10) >DroSec_CAF1 23002 113 - 1 AGUGAAGAGGCGGUGGG-UCCAAGGCGUCCAAGGGGGGUGGGUA-GUGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCCCAGUUAAGUGUGCGUGUUCAAGGAUGCAUUUUUGU ...........((((.(-(((((.(((..((....((.(((((.-(((((((((((((....)))))))))))))..))))).))...))..))).))...)))).))))..... ( -46.70) >DroEre_CAF1 24064 113 - 1 AGUGAAGAGGCGUUGGG-UCCAAGGAGUCCAAAGGGGGUGGGUU-UCGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCCCAGUUAAGUGUGCGUGUUCAAGGAUGCAUUUUUGU ..((((.(.((((((((-((....)).)))))...((.((((((-(((((((((((((....)))))))))))).))))))).)).....))).).))))............... ( -44.70) >DroYak_CAF1 25253 114 - 1 AGUGGAGAGGCGGUGGG-UCCAAGGAGUCCAAAGGGGGUGGGGAACUGCGGGUGGCAAGAGGUUGCCACUCACGCAGGCCCAGUUAAGUGUGCGUGUUCAAGGAUGCAUUUUUGU ..((((.(.(((.((((-((....)).))))....((.((((...(((((((((((((....))))))).).))))).)))).)).....))).).))))............... ( -38.40) >DroAna_CAF1 5217 110 - 1 AUCGGUGGGACAGUCUUGAAGAAAGGAUCCUGGCGGGCU----G-UCGAGGGAGGCAAGAGGCUGCCACUCACGCAGGCCCAGUUAAGUGUGCGUGUUCAAGGAUGCAUUUUUGU ...(.(((((...((((.....)))).))))).)(((((----.-.((.(((.((((......)))).))).))..)))))...((((.((((((.(....).)))))).)))). ( -35.30) >consensus AGUGGAGAGGCGGUGGG_UCCAAGGAGUCCAAGGGGGGUGGGUA_GUGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCCCAGUUAAGUGUGCGUGUUCAAGGAUGCAUUUUUGU ..((((((.((..(((...)))....((((...................(((((((((....)))))))))(((((.((........)).)))))......)))))).)))))). (-29.36 = -28.80 + -0.56)


| Location | 17,138,600 – 17,138,715 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.03 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17138600 115 + 23771897 CUGCGUGAGUGGCAACCUCUUGCCACCCACAC-UACCCAACCCCCUCGGACGCCAUGGA-ACCACCGCCUCUCCACUGCAGACAGAAAAAAAUAUUGACCAAAUGGGAAAGGAUAAC (((((((.(((((((....))))))).)))..-..............(((.((..((..-..))..))...)))...)))).............((..((....))..))....... ( -27.40) >DroSec_CAF1 23040 114 + 1 CUGCGUGAGUGGCAACCUCUUGCCACCCACAC-UACCCACCCCCCUUGGACGCCUUGGA-CCCACCGCCUCUUCACUGCAGACAGAAAAAAA-ACUGACCAAUUUCGACAGGAUAAC (((((((.(((((((....))))))).)))..-..............(((.((..((..-..))..)).))).....)))).(((.......-.))).................... ( -25.80) >DroEre_CAF1 24102 101 + 1 CUGCGUGAGUGGCAACCUCUUGCCACCCACGA-AACCCACCCCCUUUGGACUCCUUGGA-CCCAACGCCUCUUCACUACAGACAGAGAAAAG-GUUGACCAA-------------AU ...((((.(((((((....))))))).)))).-...........(((((.....(((..-..))).((((.(((.((......)).))).))-))...))))-------------). ( -27.10) >DroYak_CAF1 25291 115 + 1 CUGCGUGAGUGGCAACCUCUUGCCACCCGCAGUUCCCCACCCCCUUUGGACUCCUUGGA-CCCACCGCCUCUCCACUACACUCAGAAAAAAU-AUUUACCGAAUUGGUCAGGACAAU (((((.(.(((((((....))))))))))))).(((..(((...(((((......((((-...........))))((......)).......-.....)))))..)))..))).... ( -28.80) >DroAna_CAF1 5255 108 + 1 CUGCGUGAGUGGCAGCCUCUUGCCUCCCUCGA-C----AGCCCGCCAGGAUCCUUUCUUCAAGACUGUCCCACCGAUGCACUGAAGAAAAUU-AUUGAAAAUUUUUUACAUUAU--- .((((((((.(((((....)))))...)))((-(----((..(...((((.....))))...).)))))......))))).((.((((((((-......)))))))).))....--- ( -22.70) >consensus CUGCGUGAGUGGCAACCUCUUGCCACCCACAA_UACCCACCCCCCUUGGACGCCUUGGA_CCCACCGCCUCUCCACUGCAGACAGAAAAAAA_AUUGACCAAAUUGGACAGGAUAAC (((((((.(((((((....))))))).))).................(((.....(((...))).......)))...)))).................................... (-14.72 = -14.96 + 0.24)



| Location | 17,138,600 – 17,138,715 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.03 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -19.80 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17138600 115 - 23771897 GUUAUCCUUUCCCAUUUGGUCAAUAUUUUUUUCUGUCUGCAGUGGAGAGGCGGUGGU-UCCAUGGCGUCCGAGGGGGUUGGGUA-GUGUGGGUGGCAAGAGGUUGCCACUCACGCAG ..(((((..((((.(((((.(....(((((..(((....)))..)))))((.(((..-..))).))).)))))))))..)))))-(((((((((((((....))))))))))))).. ( -53.30) >DroSec_CAF1 23040 114 - 1 GUUAUCCUGUCGAAAUUGGUCAGU-UUUUUUUCUGUCUGCAGUGAAGAGGCGGUGGG-UCCAAGGCGUCCAAGGGGGGUGGGUA-GUGUGGGUGGCAAGAGGUUGCCACUCACGCAG .....((..((....((((.(.((-((((((.(((....))).))))))))((....-.)).....).))))....))..))..-(((((((((((((....))))))))))))).. ( -43.90) >DroEre_CAF1 24102 101 - 1 AU-------------UUGGUCAAC-CUUUUCUCUGUCUGUAGUGAAGAGGCGUUGGG-UCCAAGGAGUCCAAAGGGGGUGGGUU-UCGUGGGUGGCAAGAGGUUGCCACUCACGCAG ..-------------(((((((((-((((((.(((....))).)))))))..)))).-.))))(((..(((.......)))..)-))(((((((((((....))))))))))).... ( -38.60) >DroYak_CAF1 25291 115 - 1 AUUGUCCUGACCAAUUCGGUAAAU-AUUUUUUCUGAGUGUAGUGGAGAGGCGGUGGG-UCCAAGGAGUCCAAAGGGGGUGGGGAACUGCGGGUGGCAAGAGGUUGCCACUCACGCAG ....((((.(((.((((((.(((.-...))).))))))............(..((((-((....)).))))..)..))).)))).(((((((((((((....))))))).).))))) ( -39.90) >DroAna_CAF1 5255 108 - 1 ---AUAAUGUAAAAAAUUUUCAAU-AAUUUUCUUCAGUGCAUCGGUGGGACAGUCUUGAAGAAAGGAUCCUGGCGGGCU----G-UCGAGGGAGGCAAGAGGCUGCCACUCACGCAG ---...(((((.((((((......-))))))......)))))..(((.((((((((...((........))...)))))----)-))(((((.(((.....))).)).))).))).. ( -26.30) >consensus AUUAUCCUGUCCAAAUUGGUCAAU_AUUUUUUCUGUCUGCAGUGGAGAGGCGGUGGG_UCCAAGGAGUCCAAGGGGGGUGGGUA_GUGUGGGUGGCAAGAGGUUGCCACUCACGCAG ....................................((((......(.(((..(((...)))....)))).................(((((((((((....))))))))))))))) (-19.80 = -20.16 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:29 2006