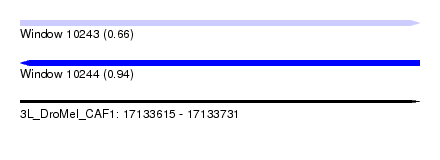
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,133,615 – 17,133,731 |
| Length | 116 |
| Max. P | 0.938020 |

| Location | 17,133,615 – 17,133,731 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.60 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17133615 116 + 23771897 ----GUGCGAAAACUGUAUAGUAAAACUGUCUGACUUUCAUAAUAGAAAAGUGGCAUUUUAAUGGGUUGGUGUCAGAACAGUUUUACUUUAUAUUCUGCCCUGACCUUGCGAUAAAUCAU ----.(((((....((((.((((((((((((((((..(((......((((......)))).......))).))))).))))))))))).)))).((......))..)))))......... ( -26.82) >DroSec_CAF1 18125 114 + 1 ----GUGCGAAAAUUGUAGAGUAAAACUGUCUGACUUUCAUA--UAAUAAGUGGCAUUUUAAUGGCUUGGUGUCAGAACAGUUUUCCUUUAUAUUCUGCCCUGACCUUGCGAUAAAUCAU ----.(((((....(((((((.(((((((((((((..(((..--((.((((......)))).))...))).))))).)))))))).))))))).((......))..)))))......... ( -26.10) >DroSim_CAF1 14892 114 + 1 ----GUGCGAAAAUUGUAGAGUAAAACUGUCUGACUUUCAUU--UAAUAAGUGGCAUUUUAAUGGCUUGGUGUCAGAACAGUUUUACUUUAUAUUCUGCCCUGACCUUGCGAUAAAUCAU ----.(((((....(((((((((((((((((((((..(((.(--((.((((......)))).)))..))).))))).)))))))))))))))).((......))..)))))......... ( -30.30) >DroEre_CAF1 19203 116 + 1 --GUGUGCGAAAAUUGCAGACUAAAACUGUCUGACUUUUAUA--UAAUAAGUGGCAUUUUAAUGGGCUUGUGUCGGAACAGUUUUAUUUUAUAUUCUACCCUGACCUUGCGAUAAAUCAU --((.(((((...))))).))((((((((((((((.......--..((((((..(((....))).))))))))))).))))))))).................................. ( -22.80) >DroYak_CAF1 20337 118 + 1 AAGUGUGCGAAAAUUGUAGAGUAAAACUGUCUGACUUUUAUA--UAAUAAGUGGCGUUUUAAUGGGUUGAUGUCAGAACAGUUUUACAUUAUAUUCUACCCUGACCUUGCGAUAAAUCAU .....(((((.....((((((((((((((((((((..(((..--((.((((......)))).))...))).))))).)))))))))).......))))).......)))))......... ( -24.91) >consensus ____GUGCGAAAAUUGUAGAGUAAAACUGUCUGACUUUCAUA__UAAUAAGUGGCAUUUUAAUGGGUUGGUGUCAGAACAGUUUUACUUUAUAUUCUGCCCUGACCUUGCGAUAAAUCAU .....(((((....(((((((((((((((((((((..((((.........))))(((....))).......))))).)))))))))))))))).((......))..)))))......... (-23.84 = -24.28 + 0.44)


| Location | 17,133,615 – 17,133,731 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.60 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17133615 116 - 23771897 AUGAUUUAUCGCAAGGUCAGGGCAGAAUAUAAAGUAAAACUGUUCUGACACCAACCCAUUAAAAUGCCACUUUUCUAUUAUGAAAGUCAGACAGUUUUACUAUACAGUUUUCGCAC---- .((((((......))))))..(((((((.((.(((((((((((.(((((.......(((....))).....((((......)))))))))))))))))))).))..))))).))..---- ( -27.80) >DroSec_CAF1 18125 114 - 1 AUGAUUUAUCGCAAGGUCAGGGCAGAAUAUAAAGGAAAACUGUUCUGACACCAAGCCAUUAAAAUGCCACUUAUUA--UAUGAAAGUCAGACAGUUUUACUCUACAAUUUUCGCAC---- .((((((......))))))..(((((((.((.((.((((((((.(((((.....(.(((....))).)..((((..--.))))..))))))))))))).)).))..))))).))..---- ( -20.00) >DroSim_CAF1 14892 114 - 1 AUGAUUUAUCGCAAGGUCAGGGCAGAAUAUAAAGUAAAACUGUUCUGACACCAAGCCAUUAAAAUGCCACUUAUUA--AAUGAAAGUCAGACAGUUUUACUCUACAAUUUUCGCAC---- .((((((......))))))..(((((((.((.(((((((((((.(((((.....(.(((....))).)..((((..--.))))..)))))))))))))))).))..))))).))..---- ( -24.60) >DroEre_CAF1 19203 116 - 1 AUGAUUUAUCGCAAGGUCAGGGUAGAAUAUAAAAUAAAACUGUUCCGACACAAGCCCAUUAAAAUGCCACUUAUUA--UAUAAAAGUCAGACAGUUUUAGUCUGCAAUUUUCGCACAC-- .((((((......))))))..(((((........((((((((((..(((..........(((........)))...--.......))).)))))))))).))))).............-- ( -17.65) >DroYak_CAF1 20337 118 - 1 AUGAUUUAUCGCAAGGUCAGGGUAGAAUAUAAUGUAAAACUGUUCUGACAUCAACCCAUUAAAACGCCACUUAUUA--UAUAAAAGUCAGACAGUUUUACUCUACAAUUUUCGCACACUU .((((((......))))))..(((((.......((((((((((.(((((..........(((........)))...--.......))))))))))))))))))))............... ( -23.36) >consensus AUGAUUUAUCGCAAGGUCAGGGCAGAAUAUAAAGUAAAACUGUUCUGACACCAACCCAUUAAAAUGCCACUUAUUA__UAUGAAAGUCAGACAGUUUUACUCUACAAUUUUCGCAC____ .((((((......))))))..(((((((.((.(((((((((((.(((((....................................)))))))))))))))).))..))))).))...... (-20.06 = -20.30 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:23 2006