
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,128,630 – 17,128,755 |
| Length | 125 |
| Max. P | 0.999081 |

| Location | 17,128,630 – 17,128,729 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
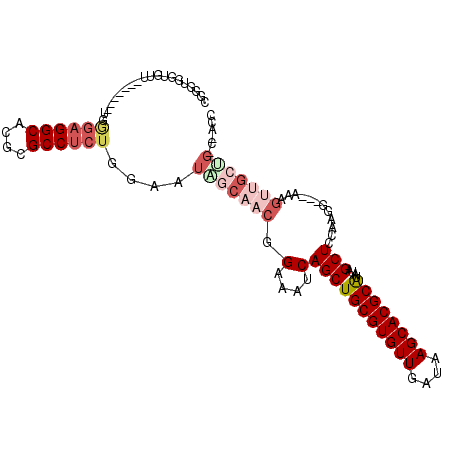
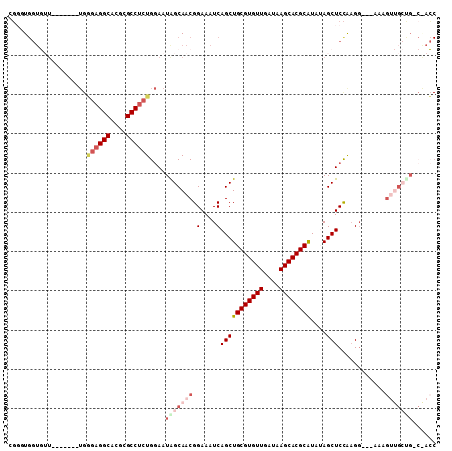
>3L_DroMel_CAF1 17128630 99 + 23771897 GAGUGGGCCUGGUAAUUUCUGG-----G-G-GGCGGUUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAU .(((...((..(......)..)-----)-(-(.(.((((.((((-------(.((((((....)))))).))))).)))).)...)).)))(((((((....))))))).... ( -40.70) >DroPse_CAF1 11737 102 + 1 GAG----ACAGUCGACUGCUGA-----GUGGGUUCGGUGGUUAUUGCAGGGUGCCAGGCACGCGCCGCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAU-- ...----..........(((((-----.....(((.((.((((((.((((((((.......))))).))).)))))).)).))).)))))((((((((....)))))))).-- ( -41.80) >DroSec_CAF1 12946 100 + 1 GAGUGGGGCUGGUAAUUUCUGG-----GGG-GGCGUUUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAU ......(((((((..(..(...-----.).-.)((((.(.((((-------(.((((((....)))))).))))).)))))...)))))))(((((((....))))))).... ( -40.30) >DroEre_CAF1 13564 99 + 1 GAGUGGGGCUGGUAAUUUCGGG-----G-G-GCGGGGUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUGGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAU ......(((((((...(((.((-----(-(-(((.(...((.((-------...)).)).).))))))).)))((....))...)))))))(((((((....))))))).... ( -39.70) >DroWil_CAF1 15274 90 + 1 GGGUGGGUUUUGAAGGUG----GAGGUG-G-GCCGGGU---------------AGAGGCACGCGCCUCUGAAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCGU-- ((.((..(((((...(((----((((((-(-(((....---------------...))).).)))))))......))..)))))..)).))(((((((....)))))))..-- ( -32.00) >consensus GAGUGGGGCUGGUAAUUUCUGG_____G_G_GCCGGGUGGUGUU_______UGGGAGGCACGCGCCUCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAU .....................................................((((((....)))))).....(((...........)))(((((((....))))))).... (-19.36 = -19.68 + 0.32)

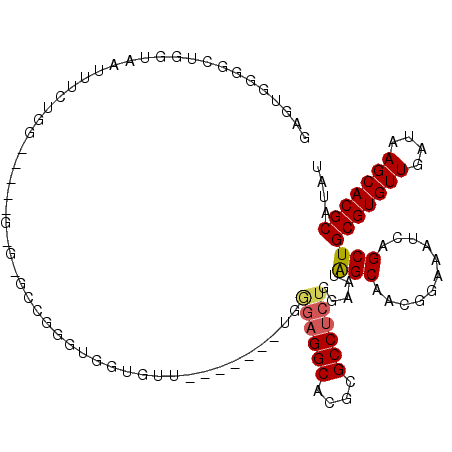
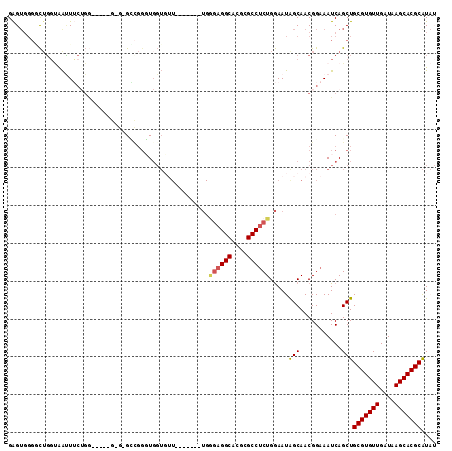
| Location | 17,128,630 – 17,128,729 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17128630 99 - 23771897 AUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGAGGCGCGUGCCUCCCA-------AACACCAACCGCC-C-C-----CCAGAAAUUACCAGGCCCACUC ...((((((........))))))(((((((((.((((........(((((....)))))...-------.....)))).(..-.-.-----.).))))))....)))...... ( -20.29) >DroPse_CAF1 11737 102 - 1 --AUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGCGGCGCGUGCCUGGCACCCUGCAAUAACCACCGAACCCAC-----UCAGCAGUCGACUGU----CUC --.((((((........))))))(((((...((((((......))))))((.(((....((.....))......))))).......-----)))))..........----... ( -27.20) >DroSec_CAF1 12946 100 - 1 AUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGAGGCGCGUGCCUCCCA-------AACACCAAACGCC-CCC-----CCAGAAAUUACCAGCCCCACUC ...((((((........))))))(((((((((((((.........(((((....)))))...-------.......))))..-...-----...)))))...))))....... ( -22.71) >DroEre_CAF1 13564 99 - 1 AUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCCAUUCCAGAGGCGCGUGCCUCCCA-------AACACCACCCCGC-C-C-----CCCGAAAUUACCAGCCCCACUC ...((((((........))))))(((((((((...((......))(((((....)))))...-------.............-.-.-----...)))))...))))....... ( -20.60) >DroWil_CAF1 15274 90 - 1 --ACGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUUCAGAGGCGCGUGCCUCU---------------ACCCGGC-C-CACCUC----CACCUUCAAAACCCACCC --..((((((((........(((((.(....).)))))........))))))))(((...---------------....)))-.-......----.................. ( -20.49) >consensus AUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGAGGCGCGUGCCUCCCA_______AACACCACCCGCC_C_C_____CCAGAAAUUACCAGCCCCACUC ...(((((((((........(((((.(....).)))))........))))))))).......................................................... (-17.03 = -17.07 + 0.04)

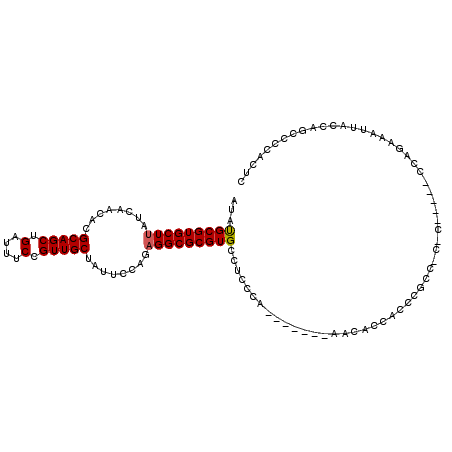
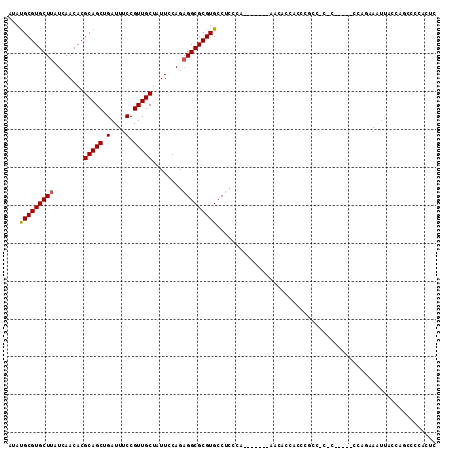
| Location | 17,128,656 – 17,128,755 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -21.98 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17128656 99 + 23771897 CGGUUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAUAGCUCCAAGG---AAAGUUGCUGCCAACC .((((((((((-------(.((((((....)))))).))))((((((........((((((((((....)))))))....)))((....)---)..))))))))))))) ( -44.30) >DroPse_CAF1 11761 92 + 1 UCGGUGGUUAUUGCAGGGUGCCAGGCACGCGCCGCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAU--AGCUCUGUGG---AG------------GC ((.((.((((((.((((((((.......))))).))).)))))).)).)).....((((((((((....)))))))).--..(((....)---))------------)) ( -35.60) >DroSec_CAF1 12973 99 + 1 CGUUUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAUAGCUCCAAGG---AAAGUUGCUGCCAACC ...((((((((-------(.((((((....)))))).))))((((((........((((((((((....)))))))....)))((....)---)..))))))))))).. ( -38.70) >DroEre_CAF1 13590 99 + 1 GGGGUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUGGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAUAGCUCCAAGG---AAAGUUGCUGGCGACC ..(((.(((((-------(.((((((....)))))).))))((((((........((((((((((....)))))))....)))((....)---)..)))))).)).))) ( -38.90) >DroWil_CAF1 15301 85 + 1 CGGGU---------------AGAGGCACGCGCCUCUGAAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCGU--AGCUCCAAGCCCAAAAGA-GACG------ .((((---------------((((((....))))))...........(((.....(((((((((........))))))--))))))..))))......-....------ ( -31.80) >DroYak_CAF1 13506 99 + 1 GGUGUGGUGUU-------UGGGAGGCACGCGCCUCUGGAAUGGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAUAGCUCCAAGG---AAAGUUGCCGACGACC ...((.(((((-------(.((((((....)))))).))))((((((........((((((((((....)))))))....)))((....)---)..)))))).)).)). ( -38.60) >consensus CGGGUGGUGUU_______UGGGAGGCACGCGCCUCUGGAAUAGCAACGGAAAUCAGCUGCGUGUUGAUAAGCACGCAUAUAGCUCCAAGG___AAAGUUGCUG_C_ACC ....................((((((....))))))....(((((((.(....)(((((((((((....))))))))....)))............)))))))...... (-21.98 = -23.87 + 1.89)

| Location | 17,128,656 – 17,128,755 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -18.87 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
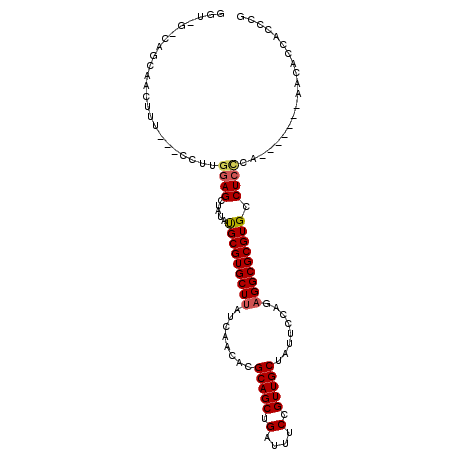
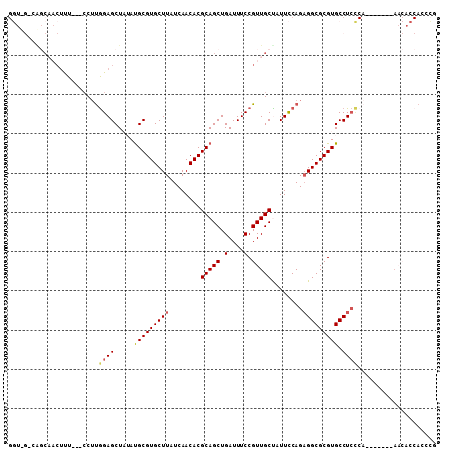
>3L_DroMel_CAF1 17128656 99 - 23771897 GGUUGGCAGCAACUUU---CCUUGGAGCUAUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGAGGCGCGUGCCUCCCA-------AACACCAACCG ((((((.((((((...---....((((.....((((((........))))))......))))))))))......(((((....)))))...-------....)))))). ( -34.71) >DroPse_CAF1 11761 92 - 1 GC------------CU---CCACAGAGCU--AUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGCGGCGCGUGCCUGGCACCCUGCAAUAACCACCGA ((------------(.---.(((..((((--..(((((........)))))))))......(((((((.....))))))).)))...)))................... ( -26.70) >DroSec_CAF1 12973 99 - 1 GGUUGGCAGCAACUUU---CCUUGGAGCUAUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGAGGCGCGUGCCUCCCA-------AACACCAAACG ((((((.((((((...---....((((.....((((((........))))))......))))))))))......(((((....))))))))-------...)))..... ( -29.41) >DroEre_CAF1 13590 99 - 1 GGUCGCCAGCAACUUU---CCUUGGAGCUAUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCCAUUCCAGAGGCGCGUGCCUCCCA-------AACACCACCCC ((((((((((....((---(....))).....((((((........)))))))))).........(((........)))))).))).....-------........... ( -26.40) >DroWil_CAF1 15301 85 - 1 ------CGUC-UCUUUUGGGCUUGGAGCU--ACGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUUCAGAGGCGCGUGCCUCU---------------ACCCG ------....-......(((..(((((..--.(((((((((........(((((.(....).)))))........))))))))).))))---------------)))). ( -28.29) >DroYak_CAF1 13506 99 - 1 GGUCGUCGGCAACUUU---CCUUGGAGCUAUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCCAUUCCAGAGGCGCGUGCCUCCCA-------AACACCACACC (((.((.((((((...---....((((.....((((((........))))))......))))))))))......(((((....)))))...-------.)))))..... ( -30.61) >consensus GGU_G_CAGCAACUUU___CCUUGGAGCUAUAUGCGUGCUUAUCAACACGCAGCUGAUUUCCGUUGCUAUUCCAGAGGCGCGUGCCUCCCA_______AACACCACCCG .......................((((.....(((((((((........(((((.(....).)))))........))))))))).)))).................... (-18.87 = -19.12 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:21 2006