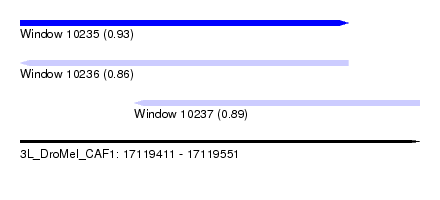
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,119,411 – 17,119,551 |
| Length | 140 |
| Max. P | 0.930865 |

| Location | 17,119,411 – 17,119,526 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -31.59 |
| Energy contribution | -31.21 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17119411 115 + 23771897 GAAAACGCUUUCGGCCCGGCCUGCCUUCCUCGAAUAUCCUUAAUCGUCCUGUCCUAUCGGUUUCCUGUGAGCCAGAACAAAAACAACCGAAAAACCGACCGGACG-GGGCCGAAAA ........(((((((((((....))...................(((((.(((...((((((...(((........))).....))))))......))).)))))-))))))))). ( -36.90) >DroSec_CAF1 3768 115 + 1 GAAAACGCUUUCGGCCCGGCCUGCCUUCCUGGAAUAUCCUUAAUCGCCCUGUCCUAUCGGUUUCCUGUGAGCCAGAACAAAAACAACCGAAAAACCGAACGGACA-GGGCCGAAAU ........((((((((((((..........((.....))......))).(((((..(((((((.(((.....)))........(....)..)))))))..)))))-))))))))). ( -34.29) >DroEre_CAF1 4335 116 + 1 GAAAACGCUUUCGGCCCGGCUUGCCUUCCUGGAAUAUCCUUAAUCGCCCUGUCCUAUCGGUUUCCUGUGAGCCAGAACAAAAACAACCGAAACCCCGACCGGACAAGGGCCGAAAA ........((((((((((((..........((.....))......))).(((((..((((((((.(((..............)))...))))))..))..))))).))))))))). ( -34.23) >DroYak_CAF1 3754 115 + 1 GAAAACGCUUUCGGCCCGGCUUGCCUUCCUGGAAUAUCCUUAAUCGCCCUGUCCUAUCGGUUUCCUGUGAGCCAGAACAAAAGCAACCGAAAA-CCGACCGGACAAGGGCCGAAAA ........((((((((((((..........((.....))......))).(((((..(((((((.(.((..((..........)).)).).)))-))))..))))).))))))))). ( -38.39) >consensus GAAAACGCUUUCGGCCCGGCCUGCCUUCCUGGAAUAUCCUUAAUCGCCCUGUCCUAUCGGUUUCCUGUGAGCCAGAACAAAAACAACCGAAAAACCGACCGGACA_GGGCCGAAAA ........(((((((((((....))........................(((((..((((((((.(((..............)))...))))..))))..))))).))))))))). (-31.59 = -31.21 + -0.38)



| Location | 17,119,411 – 17,119,526 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -43.98 |
| Consensus MFE | -38.19 |
| Energy contribution | -39.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17119411 115 - 23771897 UUUUCGGCCC-CGUCCGGUCGGUUUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGACGAUUAAGGAUAUUCGAGGAAGGCAGGCCGGGCCGAAAGCGUUUUC ((((((((((-.((((.(((((((((..(...(((........)))...)..))))))))).))))....(((.((....)).))).....((....))))))))))))....... ( -41.80) >DroSec_CAF1 3768 115 - 1 AUUUCGGCCC-UGUCCGUUCGGUUUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUAAGGAUAUUCCAGGAAGGCAGGCCGGGCCGAAAGCGUUUUC .(((((((((-(((((..((((((((..(...(((........)))...)..))))))))..))))).(((......((.....)).(.....)..))))))))))))........ ( -42.70) >DroEre_CAF1 4335 116 - 1 UUUUCGGCCCUUGUCCGGUCGGGGUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUAAGGAUAUUCCAGGAAGGCAAGCCGGGCCGAAAGCGUUUUC ((((((((((((((((.(((((..((..(...(((........)))...)..))..))))).))))))(((......((.....)).(.....)..)))))))))))))....... ( -42.70) >DroYak_CAF1 3754 115 - 1 UUUUCGGCCCUUGUCCGGUCGG-UUUUCGGUUGCUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUAAGGAUAUUCCAGGAAGGCAAGCCGGGCCGAAAGCGUUUUC ((((((((((((((((.(((((-(((((.((.(((........)))..)).)))))))))).))))))(((......((.....)).(.....)..)))))))))))))....... ( -48.70) >consensus UUUUCGGCCC_UGUCCGGUCGGUUUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUAAGGAUAUUCCAGGAAGGCAAGCCGGGCCGAAAGCGUUUUC .(((((((((..((((.(((((((((..(...(((........)))...)..))))))))).)))).........................((....)))))))))))........ (-38.19 = -39.00 + 0.81)

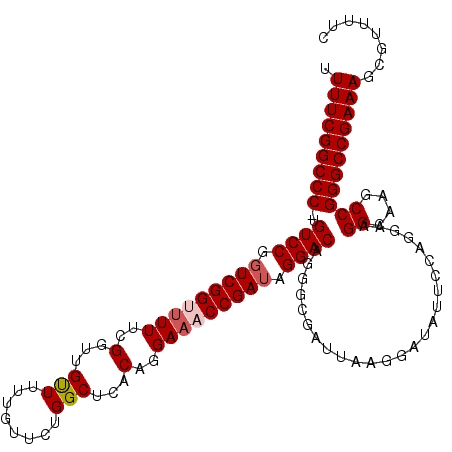
| Location | 17,119,451 – 17,119,551 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -26.56 |
| Energy contribution | -27.88 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17119451 100 - 23771897 GAUUUAUCCUUAUCCCGCCACCCAUUUUUCGGCCC-CGUCCGGUCGGUUUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGACGAUUA ...(..((((......(((...........)))..-.((((.(((((((((..(...(((........)))...)..))))))))).))))))))..)... ( -28.40) >DroSec_CAF1 3808 100 - 1 GAUUUAUCCUUAUCCUGCCACCCACAUUUCGGCCC-UGUCCGUUCGGUUUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUA ...............................((((-(((((..((((((((..(...(((........)))...)..))))))))..)))))))))..... ( -33.80) >DroEre_CAF1 4375 101 - 1 GAUUUAUUCUUAUCCUGCCUCCCAUUUUUCGGCCCUUGUCCGGUCGGGGUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUA ...............................(((((.((((.(((((..((..(...(((........)))...)..))..))))).)))))))))..... ( -29.20) >DroYak_CAF1 3794 100 - 1 GAUUUAUUCUUAUCCUGCCUCCCAUUUUUCGGCCCUUGUCCGGUCGG-UUUUCGGUUGCUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUA ...............................(((((.((((.(((((-(((((.((.(((........)))..)).)))))))))).)))))))))..... ( -35.20) >consensus GAUUUAUCCUUAUCCUGCCACCCAUUUUUCGGCCC_UGUCCGGUCGGUUUUUCGGUUGUUUUUGUUCUGGCUCACAGGAAACCGAUAGGACAGGGCGAUUA ...............................((((.(((((.(((((((((..(...(((........)))...)..))))))))).)))))))))..... (-26.56 = -27.88 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:17 2006