
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,092,154 – 17,092,248 |
| Length | 94 |
| Max. P | 0.879264 |

| Location | 17,092,154 – 17,092,248 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
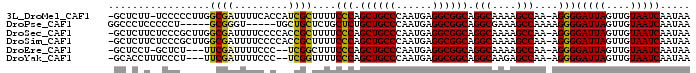
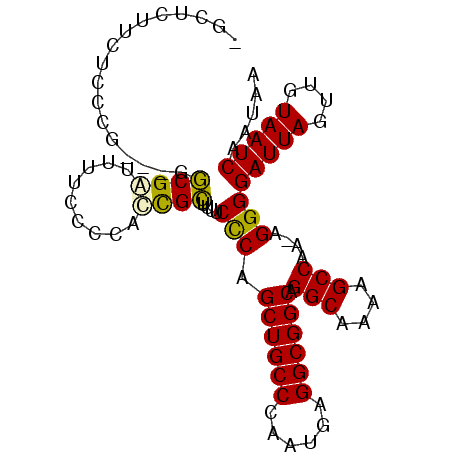
>3L_DroMel_CAF1 17092154 94 + 23771897 -GCUCUU-UCCCCCUUGGCGAUUUUCACCAUCGCUUUUCCCAGCUGCCCAAUGAGGCGGCAGGCAAAAGCCAA-AGGGGAUUAGUUGUAAUCAAUAA -(((...-(((((.((((((((.......)))..((((.((.((((((......)))))).)).)))))))))-.)))))..)))............ ( -32.50) >DroPse_CAF1 55830 87 + 1 GGCCCUCCCCCU-----GCGGGU-----UGCUGCUCUGCUCUGCUGCCCAAUGAGGCGGCAGGCGAAAGCCAAAAGGGGAUUAGUUGUAAUCAAUAA .......(((((-----((((((-----....)).))))..(((((((......)))))))(((....)))...)))))....((((....)))).. ( -34.90) >DroSec_CAF1 50304 95 + 1 -GCUCUUCUCCCGCUUGGCGAUUUUCCCCACCGCUUUUCCCAGCUGCCCAAUGAGGCGGCAGGCAAAAGCCAA-AGGGGAUUAGUUGUAAUCAAUAA -................((((((.(((((...((((((.((.((((((......)))))).)).))))))...-.)))))..))))))......... ( -33.20) >DroSim_CAF1 53234 95 + 1 -GCUCUUCUCCCGCUUGGCGAUUUUCCCCACCGCUUUUCCCAGCUGCCCAAUGAGGCGGCAGGCAAAAGCCAA-AGGGGAUUAGUUGUAAUCAAUAA -................((((((.(((((...((((((.((.((((((......)))))).)).))))))...-.)))))..))))))......... ( -33.20) >DroEre_CAF1 50346 89 + 1 -GCUCCU-GCUCU---UUCGAUUUUCCC--UCGGCUUUCCCAGCUGCCCAAUGAGGCGGCAGGCAAAAGCCAA-AGGGGAUUAGUUGUAAUCAAUAA -((....-))...---..(((((.((((--(.((((((.((.((((((......)))))).))..))))))..-.)))))..))))).......... ( -30.40) >DroYak_CAF1 53131 90 + 1 -GCACCUUUCCCU---UUCGAUUUUCCC--UCGGUUUUCCCAGCUGCCCAAUGAGGCGGCAGGCAAGAGCCAA-AGGGGAUUAGUUGUAAUCAAUAA -(((.((.(((((---((.((.......--))((((((.((.((((((......)))))).))..)))))).)-))))))..)).)))......... ( -28.50) >consensus _GCUCUUCUCCCG___GGCGAUUUUCCCCACCGCUUUUCCCAGCUGCCCAAUGAGGCGGCAGGCAAAAGCCAA_AGGGGAUUAGUUGUAAUCAAUAA .................((((.........))))....(((.((((((......)))))).(((....)))....)))(((((....)))))..... (-24.79 = -24.23 + -0.55)
| Location | 17,092,154 – 17,092,248 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -18.65 |
| Energy contribution | -19.65 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
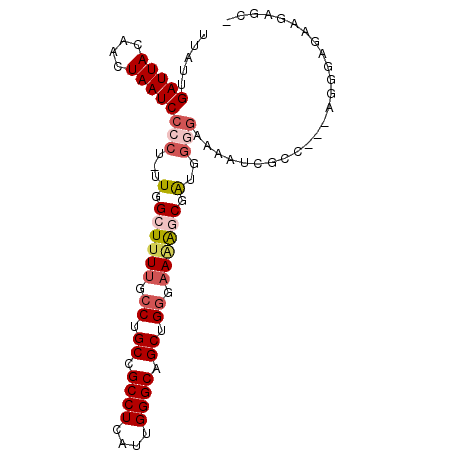
>3L_DroMel_CAF1 17092154 94 - 23771897 UUAUUGAUUACAACUAAUCCCCU-UUGGCUUUUGCCUGCCGCCUCAUUGGGCAGCUGGGAAAAGCGAUGGUGAAAAUCGCCAAGGGGGA-AAGAGC- .................((((((-((.((((((.((.((.((((....)))).)).)).))))))...((((.....))))))))))))-......- ( -37.80) >DroPse_CAF1 55830 87 - 1 UUAUUGAUUACAACUAAUCCCCUUUUGGCUUUCGCCUGCCGCCUCAUUGGGCAGCAGAGCAGAGCAGCA-----ACCCGC-----AGGGGGAGGGCC .................(((((((...(((((.((((((.((((....)))).)))).))))))).((.-----....))-----)))))))..... ( -32.80) >DroSec_CAF1 50304 95 - 1 UUAUUGAUUACAACUAAUCCCCU-UUGGCUUUUGCCUGCCGCCUCAUUGGGCAGCUGGGAAAAGCGGUGGGGAAAAUCGCCAAGCGGGAGAAGAGC- .............((..((((.(-(((((((((.((..((((((....)))).(((......)))))..)).))))..)))))).))))..))...- ( -37.80) >DroSim_CAF1 53234 95 - 1 UUAUUGAUUACAACUAAUCCCCU-UUGGCUUUUGCCUGCCGCCUCAUUGGGCAGCUGGGAAAAGCGGUGGGGAAAAUCGCCAAGCGGGAGAAGAGC- .............((..((((.(-(((((((((.((..((((((....)))).(((......)))))..)).))))..)))))).))))..))...- ( -37.80) >DroEre_CAF1 50346 89 - 1 UUAUUGAUUACAACUAAUCCCCU-UUGGCUUUUGCCUGCCGCCUCAUUGGGCAGCUGGGAAAGCCGA--GGGAAAAUCGAA---AGAGC-AGGAGC- .................((((((-(((((((((.((.((.((((....)))).)).)))))))))))--)))....((...---.))..-.)))..- ( -34.10) >DroYak_CAF1 53131 90 - 1 UUAUUGAUUACAACUAAUCCCCU-UUGGCUCUUGCCUGCCGCCUCAUUGGGCAGCUGGGAAAACCGA--GGGAAAAUCGAA---AGGGAAAGGUGC- ............(((..((((((-((((...((.((.((.((((....)))).)).)).))..))))--))).....(...---.))))..)))..- ( -26.30) >consensus UUAUUGAUUACAACUAAUCCCCU_UUGGCUUUUGCCUGCCGCCUCAUUGGGCAGCUGGGAAAAGCGAUGGGGAAAAUCGCC___AGGGAGAAGAGC_ .....(((((....)))))(((...(.((((((.((.((.((((....)))).)).)).)))))).)..)))......................... (-18.65 = -19.65 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:02 2006