| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,084,727 – 17,084,824 |
| Length | 97 |
| Max. P | 0.898245 |

| Location | 17,084,727 – 17,084,824 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.85 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.17 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17084727 97 + 23771897 -CAAGCGGUUGGCAGAGAUUAACUUUCGGGUCGCUCAAAGCGACCACAGCAAGUGCUUUACAUUUAAAACA--CGGCAAAUGGCCAA----------------AGUGCGACAG---AGA -...(((.(((((((((....((((.(.((((((.....))))))...).)))).)))).(((((......--....))))))))))----------------..))).....---... ( -27.90) >DroVir_CAF1 40342 86 + 1 GUCAGCGGUUGGCAGAGAUUAACUUUUGGGUCGCUCAAAGCGACCGCAGCAAGUGCUUUACAUUUAAAACA--CGGCGCCA----------------------------GCAG---AGG .......((((((.............((((((((.....)))))).))((..(((.((((....)))).))--).))))))----------------------------))..---... ( -27.40) >DroPse_CAF1 46853 114 + 1 -CAAGCGGUUGGCAGAGAUUAACUUUCGGGUCGCUCAAAGCGACCACAGCAAGUGCUUUACAUUUAAAACACACGGCAAUG-GGAAAAAGGCAAACACCCAACAAUGCCGCAG---AGA -.(((((.(((.(.((((.....)))).((((((.....))))))...)))).)))))...............(((((.((-((.............))))....)))))...---... ( -30.82) >DroGri_CAF1 43553 79 + 1 GUCAGCGGUUGGCAGAGAUUAACUUUUGGGUCGCUCAAAGCGACCACAGCAAGUGCUUUACAUUUAAAACA--CGGCAUCA-------------------------------------- (((....((((.(((((......)))))((((((.....)))))).))))..(((.((((....)))).))--))))....-------------------------------------- ( -22.70) >DroMoj_CAF1 49555 89 + 1 UUCAGCGGUUGGCAGAGAUUAACUUUUGGGUCGUUCAAAGCGACCACAGCAAGUGCUUUACAUUUUAAACA--UGGCGUCA----------------------------GCAGCGGAGG (((.((.((((((((((....(((((((((((((.....)))))).))).)))).)))).(((.......)--))..))))----------------------------)).)).))). ( -27.10) >DroAna_CAF1 42903 96 + 1 GCAAGCGGUUGGCAGAGAUUAACUUUCGGGUCGCUCAAAGCGACCACAGCAAGUGCUUUACAUUUAAAACA--CGGCAAUG-GGCAA----------------U-GGCCAUAG---AGA ....((.((((...((((.....)))).((((((.....)))))).))))..(((.((((....)))).))--).))....-(((..----------------.-.)))....---... ( -25.90) >consensus GCAAGCGGUUGGCAGAGAUUAACUUUCGGGUCGCUCAAAGCGACCACAGCAAGUGCUUUACAUUUAAAACA__CGGCAACA____________________________GCAG___AGA ....((.((((...((((.....)))).((((((.....)))))).))))(((((.....)))))..........)).......................................... (-18.56 = -18.17 + -0.39)

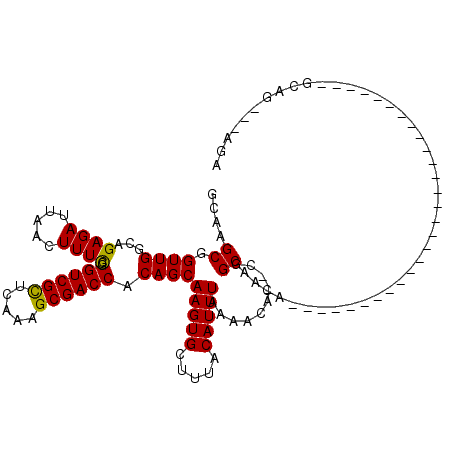

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:59 2006