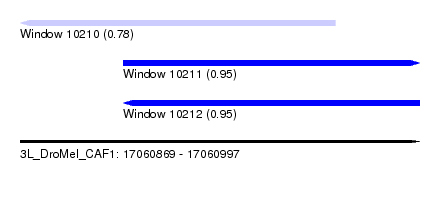
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,060,869 – 17,060,997 |
| Length | 128 |
| Max. P | 0.948307 |

| Location | 17,060,869 – 17,060,970 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -17.81 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
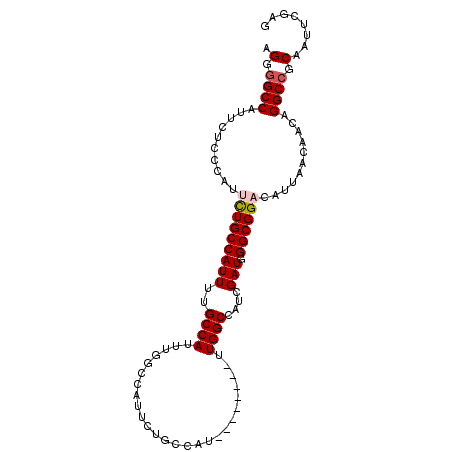
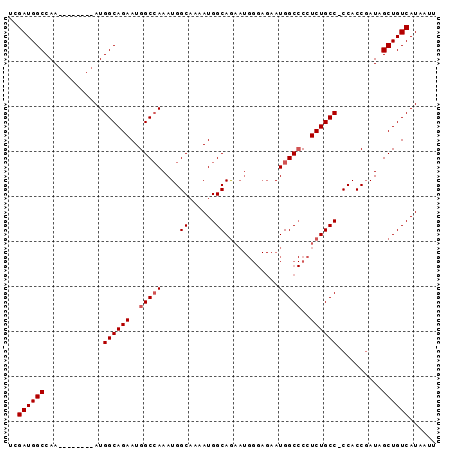
>3L_DroMel_CAF1 17060869 101 - 23771897 AGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAUUUUGCCAUUCUGCCAUUUGGCCAUUUGGCCAUCGAUGGGCGGACAUUAACAACAGGCCGCAAUUCGAG .(.((((.....((((((..((((..(.((((..(.((......)).)..)))).)..))))....)))))).(........)....)))).)........ ( -30.60) >DroSec_CAF1 19259 77 - 1 AGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAU------------------------UUGGCCAUCGAUGGGCGGACAUUAACAACAGGCCGCAAUUCGAG .(.((((..........(((((((((..(((..------------------------..)))....))).))))))...........)))).)........ ( -21.60) >DroSim_CAF1 20627 93 - 1 AGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAUUUGGCCAUUCUGCCAU--------UUGGCCAUCGAUGGGCGGACAUUAACAACAGGCCGCAAUUCGAG .(.((((..........(((((((((..((((..((((......)))).--------.))))....))).))))))...........)))).)........ ( -31.10) >DroEre_CAF1 18887 84 - 1 AGG-GCCAUU--------UUGCCAUUUGGCCAUUUGGCCAUUCUGCCAU--------UUGGCCAUCGAUGGGCGGACAUUAACAACAGGCCGCAAUUCGAG ..(-((....--------..)))...((((((..((((......)))).--------.))))))((((...((((.(..........).))))...)))). ( -32.50) >DroYak_CAF1 20322 77 - 1 AGGGGCCAUU--------UUGCCAUUUUGCCAUUUGGCC--------AU--------UUGGCAAUCGAUGGGCGGACAUUAACAACAGGCCGCAAUUCGAG .(.((((...--------((((((..(.(((....))).--------).--------.))))))..((((......)))).......)))).)........ ( -26.20) >consensus AGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAUUUGGCCAUUCUGCCAU________UUGGCCAUCGAUGGGCGGACAUUAACAACAGGCCGCAAUUCGAG .(.((((..........(((((((((..((((..........................))))....))).))))))...........)))).)........ (-17.81 = -18.17 + 0.36)

| Location | 17,060,902 – 17,060,997 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.49 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17060902 95 + 23771897 UCGAUGGCCAAAUGGCCAAAUGGCAGAAUGGCAAAAUGGCAAAAUGGCAGAAUGGGAGAAUGGCCCCUCUGCCCCCACCGAUAGCUGUCAUAAUU ....(((((....))))).(((((((....((......)).....((((((..(((........)))))))))...........))))))).... ( -31.90) >DroSim_CAF1 20660 86 + 1 UCGAUGGCCAA--------AUGGCAGAAUGGCCAAAUGGCAAAAUGGCAGAAUGGGAGAAUGGCCCCUCUGCC-CCACCUAUAGCUGUCAUAAUU ......((((.--------.((((......))))..))))...(((((((.(((((.(...(((......)))-.).)))))..))))))).... ( -28.50) >DroEre_CAF1 18920 77 + 1 UCGAUGGCCAA--------AUGGCAGAAUGGCCAAAUGGCCAAAUGGCAA--------AAUGGC-CCUCUGCC-CCACCGAUAGCUGUCAUAAUU ..((((((...--------..((((((..(((((....(((....)))..--------..))))-).))))))-.........))))))...... ( -29.54) >consensus UCGAUGGCCAA________AUGGCAGAAUGGCCAAAUGGCAAAAUGGCAGAAUGGGAGAAUGGCCCCUCUGCC_CCACCGAUAGCUGUCAUAAUU ..((((((.............((((((..(((((....((......))............)))))..))))))..........))))))...... (-19.82 = -20.49 + 0.67)

| Location | 17,060,902 – 17,060,997 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
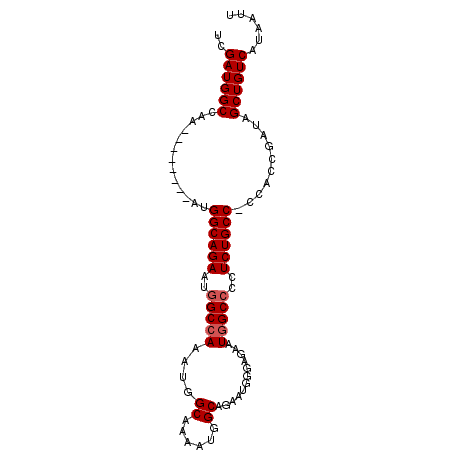
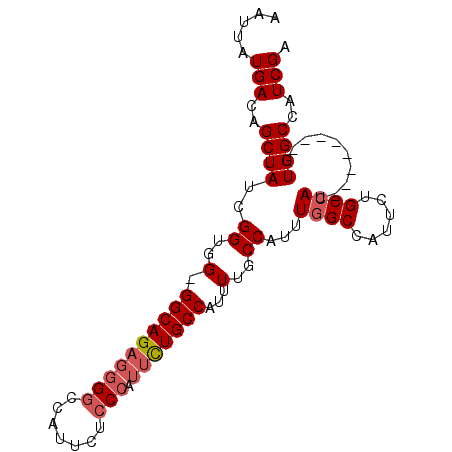
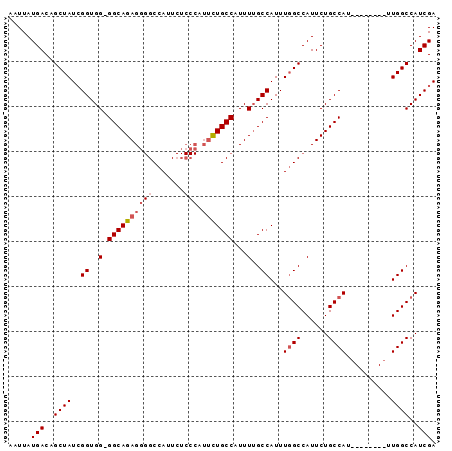
>3L_DroMel_CAF1 17060902 95 - 23771897 AAUUAUGACAGCUAUCGGUGGGGGCAGAGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAUUUUGCCAUUCUGCCAUUUGGCCAUUUGGCCAUCGA ..............((((((..((((((((((.......))).))))))).....((((..(.((((..........)))).)..)))))))))) ( -33.00) >DroSim_CAF1 20660 86 - 1 AAUUAUGACAGCUAUAGGUGG-GGCAGAGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAUUUGGCCAUUCUGCCAU--------UUGGCCAUCGA ................((((.-((((((((((.......))).))))))).....((((..((((......)))).--------.)))))))).. ( -32.20) >DroEre_CAF1 18920 77 - 1 AAUUAUGACAGCUAUCGGUGG-GGCAGAGG-GCCAUU--------UUGCCAUUUGGCCAUUUGGCCAUUCUGCCAU--------UUGGCCAUCGA ..............(((((((-((((((((-((((..--------(.(((....))).)..))))).)))))))..--------....))))))) ( -34.00) >consensus AAUUAUGACAGCUAUCGGUGG_GGCAGAGGGGCCAUUCUCCCAUUCUGCCAUUUUGCCAUUUGGCCAUUCUGCCAU________UUGGCCAUCGA .....(((..((((..((..(.((((((((((.......))).)))))))...)..))...((((......))))..........))))..))). (-20.44 = -21.97 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:50 2006