
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,060,002 – 17,060,098 |
| Length | 96 |
| Max. P | 0.990546 |

| Location | 17,060,002 – 17,060,098 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17060002 96 + 23771897 --UCUCCUGCC-UGACUGCCUGCUUAUGGGCCAUUGAUUAAGCUUAAUCAAUGCGCUUAAGGCAACUC-------GCAACUCCUGUGCACUGUAAUUAAAAUGCAG --....((((.-....(((((.....((((((((((((((....))))))))).))))))))))....-------(((.(....))))..............)))) ( -25.80) >DroVir_CAF1 13513 76 + 1 --UGUU-------------CUGCUUAUG-GCCAUUGAUUAAGUUUAAUCAAUGGGCUAAAGGCA--------------ACUUCUGUGCAUUGUAAUUAAAGUGCAG --....-------------.(((((.((-(((((((((((....)))))))).))))).)))))--------------.......((((((........)))))). ( -25.40) >DroPse_CAF1 18398 106 + 1 CGUGUCCUGCCAUUUCUGCCUGCUUAUGGGCCAUUGAUUAAGUUUAAUCAAUGCGCUUACGGCAACUCUUGCAACGCAACUCCUGUGCACAGUAAUUAAAAUGCAG ......((((.((((.(((.(((...((((((((((((((....))))))))).))))).(....)....)))..)))....(((....)))......)))))))) ( -25.00) >DroGri_CAF1 18064 76 + 1 --GAUC-------------AUGCUUAUG-GCCAUUGAUUAAAUUUAAUCAAUGGGCUUAAGGCA--------------ACUCCUGUGCAUUGUAAUUAAAGUGCAG --....-------------.(((((..(-.((((((((((....)))))))))).)...)))))--------------.......((((((........)))))). ( -24.40) >DroMoj_CAF1 19683 80 + 1 --UGUU---------CGCGCUGCUUAUG-GCCAUUGAUUAAGUUUAAUCAAUGAGCUUAAGGCA--------------ACUUCUGUGCAUUGUAAUUAAAGUGCAG --....---------.(((((.((((.(-(((((((((((....))))))))).)))))))(((--------------.(....))))...........))))).. ( -23.50) >DroAna_CAF1 18323 96 + 1 --CCUCUUGCC-UGACUGCCUGCUUAUGGGCCAUUGAUUAAACUUAAUCAAUACGCUUAAGGCAACUC-------UCAACUCCUGUGCACUGUAAUUAAAAUGCAG --....(((((-(....((((......)))).((((((((....)))))))).......))))))...-------..............(((((.......))))) ( -20.30) >consensus __UGUC_________CUGCCUGCUUAUG_GCCAUUGAUUAAGUUUAAUCAAUGCGCUUAAGGCA______________ACUCCUGUGCACUGUAAUUAAAAUGCAG ....................(((((....(((((((((((....))))))))).))...))))).....................((((.(........).)))). (-14.77 = -15.27 + 0.50)

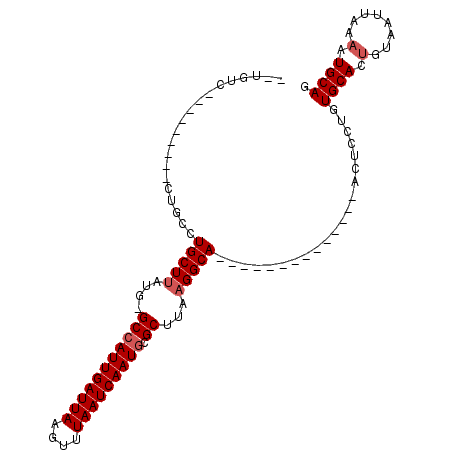
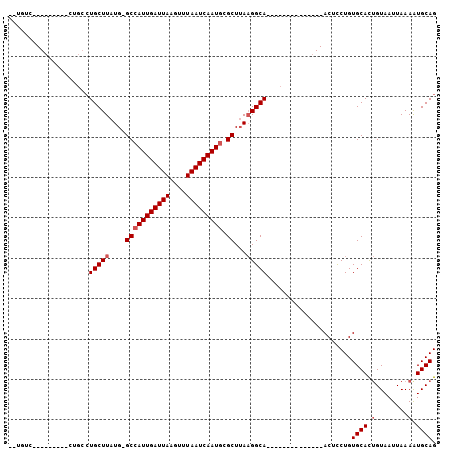
| Location | 17,060,002 – 17,060,098 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17060002 96 - 23771897 CUGCAUUUUAAUUACAGUGCACAGGAGUUGC-------GAGUUGCCUUAAGCGCAUUGAUUAAGCUUAAUCAAUGGCCCAUAAGCAGGCAGUCA-GGCAGGAGA-- ((((..............(((.......)))-------((.((((((......(((((((((....)))))))))((......)))))))))).-.))))....-- ( -25.10) >DroVir_CAF1 13513 76 - 1 CUGCACUUUAAUUACAAUGCACAGAAGU--------------UGCCUUUAGCCCAUUGAUUAAACUUAAUCAAUGGC-CAUAAGCAG-------------AACA-- ((((..............(((.......--------------))).....(.((((((((((....)))))))))).-)....))))-------------....-- ( -18.00) >DroPse_CAF1 18398 106 - 1 CUGCAUUUUAAUUACUGUGCACAGGAGUUGCGUUGCAAGAGUUGCCGUAAGCGCAUUGAUUAAACUUAAUCAAUGGCCCAUAAGCAGGCAGAAAUGGCAGGACACG .((((...(((((.(((....))).)))))...))))....(((((((.....(((((((((....)))))))))(((........)))....)))))))...... ( -29.10) >DroGri_CAF1 18064 76 - 1 CUGCACUUUAAUUACAAUGCACAGGAGU--------------UGCCUUAAGCCCAUUGAUUAAAUUUAAUCAAUGGC-CAUAAGCAU-------------GAUC-- ................((((..(((...--------------..)))...(.((((((((((....)))))))))).-)....))))-------------....-- ( -16.80) >DroMoj_CAF1 19683 80 - 1 CUGCACUUUAAUUACAAUGCACAGAAGU--------------UGCCUUAAGCUCAUUGAUUAAACUUAAUCAAUGGC-CAUAAGCAGCGCG---------AACA-- .((((............)))).....((--------------(((.(((.((.(((((((((....)))))))))))-..))))))))...---------....-- ( -18.70) >DroAna_CAF1 18323 96 - 1 CUGCAUUUUAAUUACAGUGCACAGGAGUUGA-------GAGUUGCCUUAAGCGUAUUGAUUAAGUUUAAUCAAUGGCCCAUAAGCAGGCAGUCA-GGCAAGAGG-- .((((((........))))))..........-------...((((((...((.(((((((((....)))))))))(((........))).)).)-)))))....-- ( -22.00) >consensus CUGCACUUUAAUUACAAUGCACAGGAGU______________UGCCUUAAGCGCAUUGAUUAAACUUAAUCAAUGGC_CAUAAGCAGGCAG_________GACA__ ((((..............(((.....................))).....((.(((((((((....)))))))))))......))))................... (-13.97 = -14.00 + 0.03)

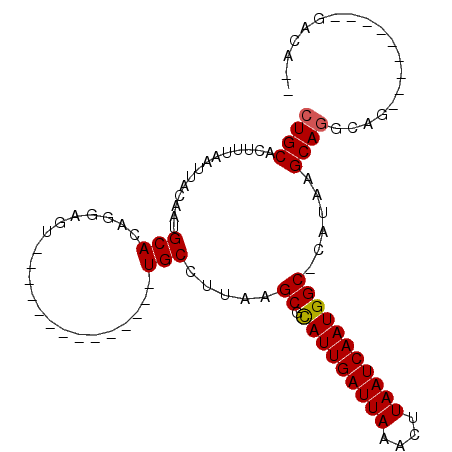
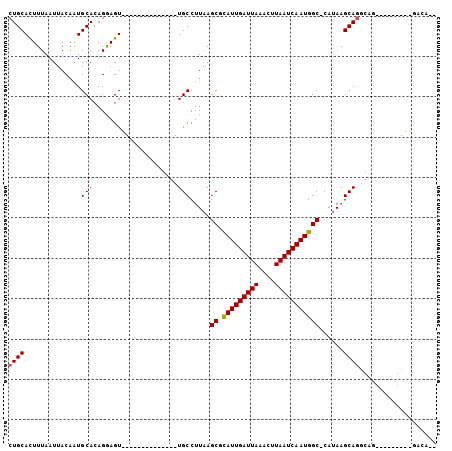
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:48 2006