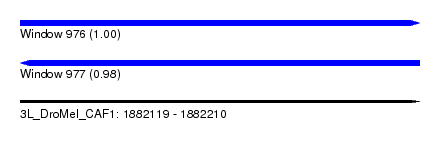
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,882,119 – 1,882,210 |
| Length | 91 |
| Max. P | 0.998191 |

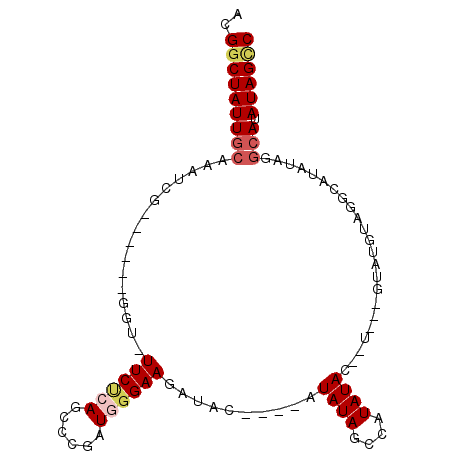
| Location | 1,882,119 – 1,882,210 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -9.52 |
| Energy contribution | -10.78 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.35 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
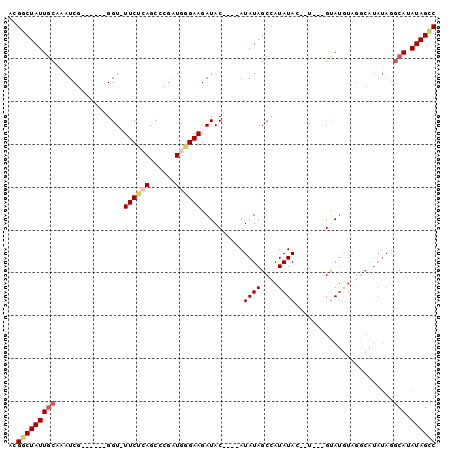
>3L_DroMel_CAF1 1882119 91 + 23771897 ACGGCUAUUGCAAAUCG------GGU-UUCUCAGCCCGAUGGGAAGAUACAUACAUAUAGCAAUAUA--UUCAGGUAUGUAGGCAUAUAGGCAUAUAGUC ..(((((((((..((((------(((-......))))))).....(.((((((((((((....))))--)....)))))))..)......))).)))))) ( -27.80) >DroSec_CAF1 4655 82 + 1 ACGGCUAUUGCGAAUCG------GGUUUUCUCAGCCCGAUGGGAAGAUAC----AUAUAGCCAUAUACAAUGUAGUAUGUAGGC--------AUAUAGCC ..((((((..(..((((------((((.....))))))))..).......----.....(((((((((......)))))).)))--------..)))))) ( -27.40) >DroSim_CAF1 4680 90 + 1 ACGGCUAUUGCGAAUCG------GGUUUUCUCAGCCCGAUGCGAAGAUAC----AUAUAGCCAUAUACAUUGUAGUAUGUAGGCAUAUAGGCAUAUAGCC ..((((((((((.((((------((((.....)))))))).)........----.(((((((((((((......)))))).))).)))).))).)))))) ( -32.90) >DroEre_CAF1 4565 88 + 1 ACGCCUAUUGCAAAUCGAGCGAGGGU-UUCCGAGCGCCAUGGGAAGAUAC----AUAUAGCCAUAUAG------GCAUAU-GCCAUAUAGGCAUAUAGCC ..((((((.((.....(.(((.((..-..))...))))(((........)----))...))...))))------))((((-(((.....))))))).... ( -26.10) >DroYak_CAF1 4699 75 + 1 ACGGCUAUUGCAAAUCG------GGU-UUCUGAGCGCGCUGGGAAGAUAC----AUAUAGCCAUAUAG------GCA--------UAUAGGCAUAUAGCC ..(((((((((.....(------..(-((((.((....)).)))))...)----.(((((((.....)------)).--------)))).))).)))))) ( -22.90) >consensus ACGGCUAUUGCAAAUCG______GGU_UUCUCAGCCCGAUGGGAAGAUAC____AUAUAGCCAUAUAC__U___GUAUGUAGGCAUAUAGGCAUAUAGCC ..(((((((((................((((((......))))))..........((((....)))).......................))).)))))) ( -9.52 = -10.78 + 1.26)

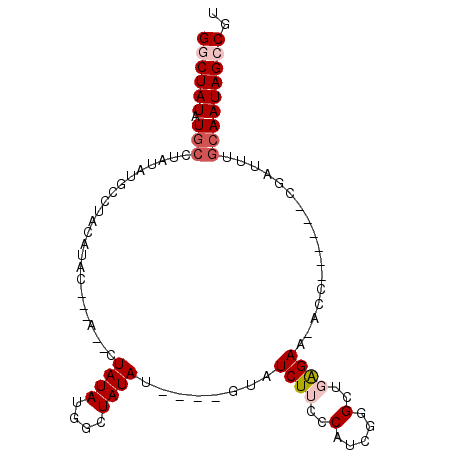
| Location | 1,882,119 – 1,882,210 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -4.62 |
| Energy contribution | -5.66 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.19 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1882119 91 - 23771897 GACUAUAUGCCUAUAUGCCUACAUACCUGAA--UAUAUUGCUAUAUGUAUGUAUCUUCCCAUCGGGCUGAGAA-ACC------CGAUUUGCAAUAGCCGU ......(((.((((.(((.(((((((....(--((((....)))))))))))).......((((((.......-.))------))))..))))))).))) ( -19.10) >DroSec_CAF1 4655 82 - 1 GGCUAUAU--------GCCUACAUACUACAUUGUAUAUGGCUAUAU----GUAUCUUCCCAUCGGGCUGAGAAAACC------CGAUUCGCAAUAGCCGU ((((((.(--------((.((((((...(((.....)))....)))----))).......((((((.........))------))))..))))))))).. ( -23.40) >DroSim_CAF1 4680 90 - 1 GGCUAUAUGCCUAUAUGCCUACAUACUACAAUGUAUAUGGCUAUAU----GUAUCUUCGCAUCGGGCUGAGAAAACC------CGAUUCGCAAUAGCCGU ((((((((((.((((.(((...((((......))))..))))))).----))))....((((((((.........))------))))..)).)))))).. ( -29.00) >DroEre_CAF1 4565 88 - 1 GGCUAUAUGCCUAUAUGGC-AUAUGC------CUAUAUGGCUAUAU----GUAUCUUCCCAUGGCGCUCGGAA-ACCCUCGCUCGAUUUGCAAUAGGCGU (((.(((((((.....)))-))))))------)...(((.((((.(----(((...((....((((...(...-.)...)))).))..)))))))).))) ( -28.50) >DroYak_CAF1 4699 75 - 1 GGCUAUAUGCCUAUA--------UGC------CUAUAUGGCUAUAU----GUAUCUUCCCAGCGCGCUCAGAA-ACC------CGAUUUGCAAUAGCCGU ((((((((((.((((--------.((------(.....))))))).----)))).........(((.((....-...------.))..))).)))))).. ( -18.60) >consensus GGCUAUAUGCCUAUAUGCCUACAUAC___A__CUAUAUGGCUAUAU____GUAUCUUCCCAUCGGGCUGAGAA_ACC______CGAUUUGCAAUAGCCGU ((((((.(((.......................((((....))))........((((..(.....)..)))).................))))))))).. ( -4.62 = -5.66 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:20 2006