
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,043,892 – 17,043,984 |
| Length | 92 |
| Max. P | 0.940607 |

| Location | 17,043,892 – 17,043,984 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -20.57 |
| Energy contribution | -22.18 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
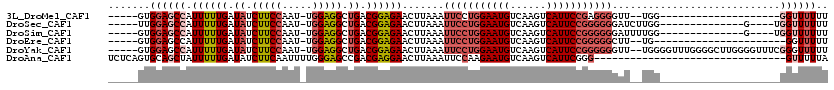
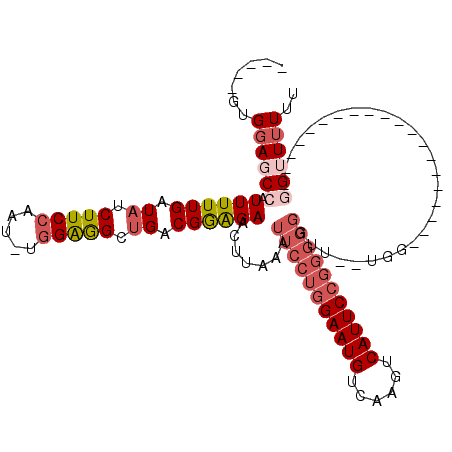
>3L_DroMel_CAF1 17043892 92 + 23771897 -----GUGGAGCCAUUUUUGAUAUCUUCCAAU-UGGAGGCUGACGGAGAACUUAAAUUCCUGGAAUGUCAAGUCAUUCCGAGGGGUU--UGG--------------------GGUUUUUU -----..((((((.((((((.((.(((((...-.))))).)).))))))..((((((((((((((((......)))))).)))))))--)))--------------------)))))).. ( -30.90) >DroSec_CAF1 11527 96 + 1 -----UUGGAGCCAUUUUUGAUAUCUUCCAAU-UGGAGGCUGACGGAGAACUUAAAUUCCUGGAAUGUCAAGUCAUUCCGGGGGGAUCUUGG--------------G----UGGUUUUUU -----..((((((.((((((.((.(((((...-.))))).)).))))))((((((.(((((((((((......)))))))))))....))))--------------)----))))))).. ( -32.50) >DroSim_CAF1 12571 96 + 1 -----GUGGAGCCAUUUUUGAUAUCUUCCAAU-UGGAGGCUGACGGAGAACUUAAAUUCCUGGAAUGUCAAGUCAUUCCGGGGGGAUUUUGG--------------G----UGGUUUUUU -----..((((((.((((((.((.(((((...-.))))).)).))))))((((((((((((((((((......)))))))))))...)))))--------------)----))))))).. ( -32.80) >DroEre_CAF1 11227 90 + 1 -----GUGGAGCCAUUUUUGAUAUCUUCCAAU-UGGAGGCUGACGGAGAACUUAAAUUCCUGGAAUGUCAAGUCAUUCCGGGGGCUU--UG----------------------GGUUUUU -----..((((((.((((((.((.(((((...-.))))).)).)))))).........(((((((((......))))))))))))))--).----------------------....... ( -31.30) >DroYak_CAF1 12448 112 + 1 -----GUGGAGCCAUUUUUGAUAUCUUCCAAU-UGGAGGCUGACGGAGAACUUAAAUUCCUGGAAUGUCAAGUCAUUCCGGGGGGUU--UGGGGUUUGGGGCUUGGGGUUUCGGGUUUUU -----..((((((.((((((.((.(((((...-.))))).)).)))))).(((((((((((((((((......)))))).)))))))--))))...((..((.....))..)))))))). ( -37.40) >DroAna_CAF1 10552 88 + 1 UCUCAGUGCAGCUAUUUUUGAUAUCUUCAAUUUUGGGAGCCGACGAGGAACUUAAAUUCCAAGAAUGUCAAGUCAUUCGGG--------------------------------GUUUUUA ((((.(.((..(((...((((.....))))...)))..)))...))))....((((..((..(((((......)))))..)--------------------------------)..)))) ( -18.00) >consensus _____GUGGAGCCAUUUUUGAUAUCUUCCAAU_UGGAGGCUGACGGAGAACUUAAAUUCCUGGAAUGUCAAGUCAUUCCGGGGGGUU__UGG____________________GGUUUUUU .......((((((.((((((.((.(((((.....))))).)).)))))).......(((((((((((......)))))))))))............................)))))).. (-20.57 = -22.18 + 1.61)
| Location | 17,043,892 – 17,043,984 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -16.65 |
| Consensus MFE | -12.97 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17043892 92 - 23771897 AAAAAACC--------------------CCA--AACCCCUCGGAAUGACUUGACAUUCCAGGAAUUUAAGUUCUCCGUCAGCCUCCA-AUUGGAAGAUAUCAAAAAUGGCUCCAC----- ........--------------------...--....(((.((((((......))))))))).................(((((((.-...))).(....)......))))....----- ( -16.60) >DroSec_CAF1 11527 96 - 1 AAAAAACCA----C--------------CCAAGAUCCCCCCGGAAUGACUUGACAUUCCAGGAAUUUAAGUUCUCCGUCAGCCUCCA-AUUGGAAGAUAUCAAAAAUGGCUCCAA----- .........----.--------------....(((......((((((......)))))).(((((....)))))..)))(((((((.-...))).(....)......))))....----- ( -16.20) >DroSim_CAF1 12571 96 - 1 AAAAAACCA----C--------------CCAAAAUCCCCCCGGAAUGACUUGACAUUCCAGGAAUUUAAGUUCUCCGUCAGCCUCCA-AUUGGAAGAUAUCAAAAAUGGCUCCAC----- .........----.--------------............((((..(((((((.((((...))))))))))).))))..(((((((.-...))).(....)......))))....----- ( -15.90) >DroEre_CAF1 11227 90 - 1 AAAAACC----------------------CA--AAGCCCCCGGAAUGACUUGACAUUCCAGGAAUUUAAGUUCUCCGUCAGCCUCCA-AUUGGAAGAUAUCAAAAAUGGCUCCAC----- .......----------------------..--.((((((.((((((......)))))).)).......((..((((..........-..))))..)).........))))....----- ( -18.30) >DroYak_CAF1 12448 112 - 1 AAAAACCCGAAACCCCAAGCCCCAAACCCCA--AACCCCCCGGAAUGACUUGACAUUCCAGGAAUUUAAGUUCUCCGUCAGCCUCCA-AUUGGAAGAUAUCAAAAAUGGCUCCAC----- .................((((..........--.....((.((((((......)))))).)).......((..((((..........-..))))..)).........))))....----- ( -18.30) >DroAna_CAF1 10552 88 - 1 UAAAAAC--------------------------------CCCGAAUGACUUGACAUUCUUGGAAUUUAAGUUCCUCGUCGGCUCCCAAAAUUGAAGAUAUCAAAAAUAGCUGCACUGAGA ......(--------------------------------(..(((((......)))))..))...........((((((((((.......((((.....))))....))))).)).))). ( -14.60) >consensus AAAAAACC____________________CCA__AACCCCCCGGAAUGACUUGACAUUCCAGGAAUUUAAGUUCUCCGUCAGCCUCCA_AUUGGAAGAUAUCAAAAAUGGCUCCAC_____ .........................................((((((......)))))).(((((....))))).....(((((((.....))).(....)......))))......... (-12.97 = -12.92 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:43 2006