
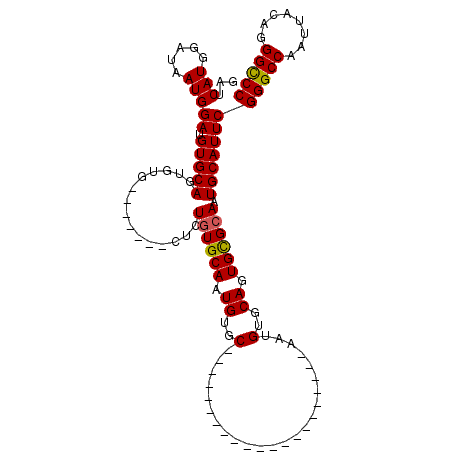
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,040,902 – 17,040,999 |
| Length | 97 |
| Max. P | 0.996001 |

| Location | 17,040,902 – 17,040,999 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.73 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17040902 97 + 23771897 GGGCCCUGUAAUUGGCCCGAAUGCAUUGCACACUGCACAUUGUCCAGUGCAAACU-------GCACAUUGCACAUUG-------CACAUUGCACAUCCAUUAUCCAUGAUC (((((........)))))...((((.((((...((((...(((.((((....)))-------).))).))))...))-------))...)))).................. ( -28.50) >DroSec_CAF1 8547 83 + 1 GGACCCUGUAAUUGGCCCGAAUGCAUUGCGCACUGCACAUU---------------------GCACAUUGCACAGAG-------CACACUGCACAUCCAUUAUCCAUGAUC (((...((((....((.(...((((.((.(((........)---------------------)).)).))))..).)-------)....))))..)))............. ( -17.60) >DroSim_CAF1 9573 83 + 1 GGGCCCUGUAAUUGGCCCGAAUGCAUUGCGCACUGCACAUU---------------------GCACAUUGCACAGAG-------CACACUGCACAUCCAUUAUCCAUGAUC (((((........)))))...((((.((.((.(((.....(---------------------((.....)))))).)-------).)).)))).................. ( -23.30) >DroYak_CAF1 9403 111 + 1 GGGCCCUGUAAUUGGCCCGAAUGCAUUGCGCAUUGCACACAGCACACAGCACACUGCACAUUGCACAUUGCAGAGGGCACACUGCACACUGCACAUCCAUUAUCCAUGAUC (((((........)))))...((((.((.((..(((.....)))....)).)).))))...((((...(((((.(....).)))))...)))).................. ( -30.50) >consensus GGGCCCUGUAAUUGGCCCGAAUGCAUUGCGCACUGCACAUU_____________________GCACAUUGCACAGAG_______CACACUGCACAUCCAUUAUCCAUGAUC (((((........)))))...((((.(((.....))).........................((.....))..................)))).................. (-14.48 = -14.73 + 0.25)

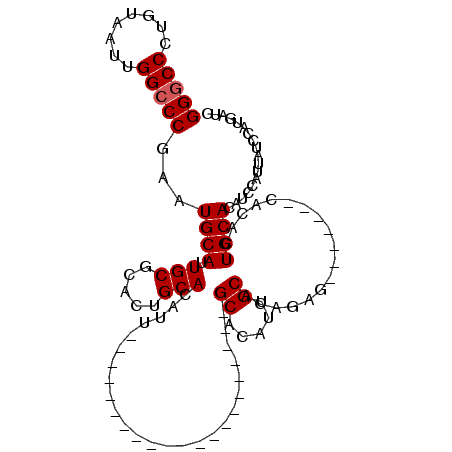

| Location | 17,040,902 – 17,040,999 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.45 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17040902 97 - 23771897 GAUCAUGGAUAAUGGAUGUGCAAUGUG-------CAAUGUGCAAUGUGC-------AGUUUGCACUGGACAAUGUGCAGUGUGCAAUGCAUUCGGGCCAAUUACAGGGCCC ...(((.....)))((.(((((.((..-------((.((..((.(((.(-------(((....)))).))).))..)).))..)).)))))))(((((........))))) ( -39.00) >DroSec_CAF1 8547 83 - 1 GAUCAUGGAUAAUGGAUGUGCAGUGUG-------CUCUGUGCAAUGUGC---------------------AAUGUGCAGUGCGCAAUGCAUUCGGGCCAAUUACAGGGUCC ...(((.....)))((.(((((.((((-------(.(((..((......---------------------..))..))).))))).)))))))(((((........))))) ( -30.90) >DroSim_CAF1 9573 83 - 1 GAUCAUGGAUAAUGGAUGUGCAGUGUG-------CUCUGUGCAAUGUGC---------------------AAUGUGCAGUGCGCAAUGCAUUCGGGCCAAUUACAGGGCCC ...(((.....)))((.(((((.((((-------(.(((..((......---------------------..))..))).))))).)))))))(((((........))))) ( -33.60) >DroYak_CAF1 9403 111 - 1 GAUCAUGGAUAAUGGAUGUGCAGUGUGCAGUGUGCCCUCUGCAAUGUGCAAUGUGCAGUGUGCUGUGUGCUGUGUGCAAUGCGCAAUGCAUUCGGGCCAAUUACAGGGCCC .............((((((((((.(.((.....)).).))))..((((((.((..((..(..(...)..)..))..)).))))))..))))))(((((........))))) ( -41.30) >consensus GAUCAUGGAUAAUGGAUGUGCAGUGUG_______CUCUGUGCAAUGUGC_____________________AAUGUGCAGUGCGCAAUGCAUUCGGGCCAAUUACAGGGCCC ...(((.....)))((.(((((...............((((((.((..(........................)..)).)))))).)))))))(((((........))))) (-20.58 = -20.45 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:40 2006