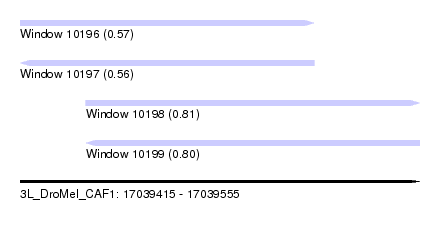
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,039,415 – 17,039,555 |
| Length | 140 |
| Max. P | 0.806262 |

| Location | 17,039,415 – 17,039,518 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.07 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -18.63 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17039415 103 + 23771897 UGUGGUUGUGUCUAUUUG-----------GCAAUGAGCCGCACUUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAAGCAGCAUA ..((((((((..((((((-----------((.....))).......((....))...((((.((((.......)))).)))))))))..))))))))((.((....)).))... ( -37.20) >DroVir_CAF1 1578 114 + 1 UGGAGGUGUGUCUGUGAGAGAGUAGAAGAGCAGUUAAACAAACUUACCAGUUGGAUAGGGUAGUCCGAAGCUCGGAUAGCCGGAGUAGCCGCGACCUGCGGCAUAGGCAGCAUA .....(((((((((((.....((((....((.((((..(.......((....))....(((.(((((.....))))).))))...)))).))...))))..)))))))).))). ( -34.60) >DroPse_CAF1 7030 87 + 1 UA--------------UG-----------G--GUGAACGGGACUUACCAGUUGGAUAGGGGAGUCCGAAGGAGGGAUAGCCCGAAUAUCCGCGACCAGCGGCAUAGGCAGCAUA ..--------------.(-----------(--(((..((((.((..((..((((((......))))))....))...))))))..)))))((..((.........))..))... ( -26.10) >DroMoj_CAF1 7343 98 + 1 AGAGGGAGAGU--GAGAUAGAGU---------GUU-----GACUUACCAGUUGGAUAGGGUAGUCCGAAGCUGGGAUAACCCGAAUAGCCGCGACCAGCGGCAUAAGCAGCAUA ...........--........((---------(((-----(.((((((....))...((((.((((.......)))).)))).....(((((.....))))).)))))))))). ( -32.30) >DroAna_CAF1 6134 91 + 1 UGUGU------------G-----------GGUGUGCGUCGGACUUACCAGUUGGAUAGGGCAGACCGAAGCUGGGAUAGCCCGAGUAGCCACGACCAGCGGCAUAAGCAGCAUA (((((------------.-----------(((.((((((.((((....)))).)))...))).)))...((((((....))).....(((.(.....).)))...))).))))) ( -29.60) >DroPer_CAF1 697 87 + 1 UA--------------UG-----------G--GUGAACGGGACUUACCAGUUGGAUAGGGGAGUCCGAAGGAGGGAUAGCCCGAAUAUCCGCGACCAGCGGCAUAGGCAGCAUA ..--------------.(-----------(--(((..((((.((..((..((((((......))))))....))...))))))..)))))((..((.........))..))... ( -26.10) >consensus UG_GG___________UG___________G__GUGAACCGGACUUACCAGUUGGAUAGGGCAGUCCGAAGCUGGGAUAGCCCGAAUAGCCGCGACCAGCGGCAUAAGCAGCAUA .................................................((((....((((.((((.......)))).))))......((((.....))))......))))... (-18.63 = -19.27 + 0.64)


| Location | 17,039,415 – 17,039,518 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.07 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -14.97 |
| Energy contribution | -14.83 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17039415 103 - 23771897 UAUGCUGCUUAUGCCGCUGGUCGCGGAUACUCGGGCUAUCCCAGUUUUGGACUGCCCUAUCCAACUGGUAAGUGCGGCUCAUUGC-----------CAAAUAGACACAACCACA ...((.((....)).))((((((((((((...((((..(((.......)))..))))))))).(((....)))))(((.....))-----------).....))).))...... ( -32.20) >DroVir_CAF1 1578 114 - 1 UAUGCUGCCUAUGCCGCAGGUCGCGGCUACUCCGGCUAUCCGAGCUUCGGACUACCCUAUCCAACUGGUAAGUUUGUUUAACUGCUCUUCUACUCUCUCACAGACACACCUCCA ...((((.....(((((.....))))).....)))).....((((..((((((..((.........))..)))))).......))))........................... ( -25.50) >DroPse_CAF1 7030 87 - 1 UAUGCUGCCUAUGCCGCUGGUCGCGGAUAUUCGGGCUAUCCCUCCUUCGGACUCCCCUAUCCAACUGGUAAGUCCCGUUCAC--C-----------CA--------------UA ((((..(((((((((((.....)))).)))..))))............(((((..((.........))..))))).......--.-----------))--------------)) ( -21.60) >DroMoj_CAF1 7343 98 - 1 UAUGCUGCUUAUGCCGCUGGUCGCGGCUAUUCGGGUUAUCCCAGCUUCGGACUACCCUAUCCAACUGGUAAGUC-----AAC---------ACUCUAUCUC--ACUCUCCCUCU ..((.((((((((((((.....))))).....((((..(((.......)))..))))..........))))).)-----).)---------).........--........... ( -23.50) >DroAna_CAF1 6134 91 - 1 UAUGCUGCUUAUGCCGCUGGUCGUGGCUACUCGGGCUAUCCCAGCUUCGGUCUGCCCUAUCCAACUGGUAAGUCCGACGCACACC-----------C------------ACACA ..((.(((.......(((((..((((((.....)))))).))))).((((.((..((.........))..)).)))).)))))..-----------.------------..... ( -27.00) >DroPer_CAF1 697 87 - 1 UAUGCUGCCUAUGCCGCUGGUCGCGGAUAUUCGGGCUAUCCCUCCUUCGGACUCCCCUAUCCAACUGGUAAGUCCCGUUCAC--C-----------CA--------------UA ((((..(((((((((((.....)))).)))..))))............(((((..((.........))..))))).......--.-----------))--------------)) ( -21.60) >consensus UAUGCUGCCUAUGCCGCUGGUCGCGGAUACUCGGGCUAUCCCAGCUUCGGACUACCCUAUCCAACUGGUAAGUCCGGUUCAC__C___________CA___________CC_CA .............((((.....))))......(((....)))......(((((..((.........))..)))))....................................... (-14.97 = -14.83 + -0.13)
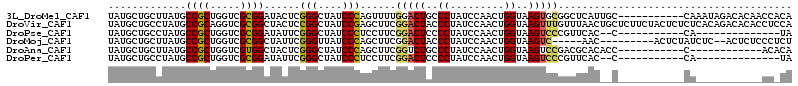
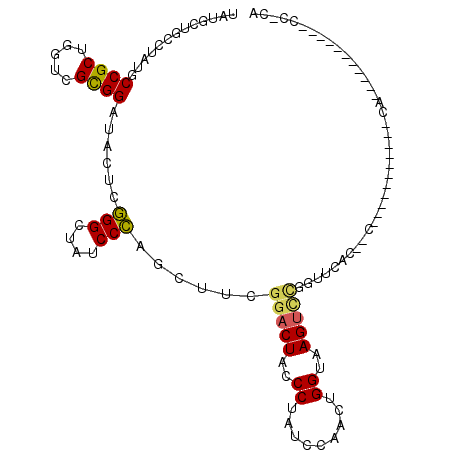
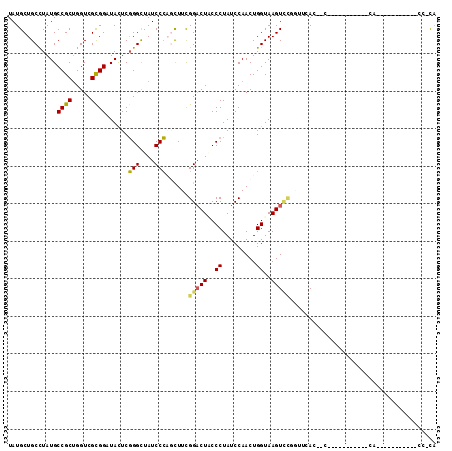
| Location | 17,039,438 – 17,039,555 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17039438 117 + 23771897 GAGCCGCACUUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAAGCAGCAUAGGCAGCUGCCGGAAAACCUAAUCGAAAAAG---UAUAGAA ..((((((((....))).(((((((((.((((.......)))).))))...))))).......)))))....(((((.......))))).((....))............---....... ( -35.90) >DroVir_CAF1 1612 114 + 1 UAAACAAACUUACCAGUUGGAUAGGGUAGUCCGAAGCUCGGAUAGCCGGAGUAGCCGCGACCUGCGGCAUAGGCAGCAUACGCAGCUGCCGGAAAGCCUGCGAGAAAGAA-----AAC-A ........(((.((....))...((((..(((...((((((....)).)))).(((((.....)))))...((((((.......)))))))))..))))..)))......-----...-. ( -38.40) >DroGri_CAF1 7097 113 + 1 UGAGC-AACUUACCAGUUGGAUAGGGUAGUCCAAAGCUAGGAUAGCCGGAGUAACCGCGACCAGAGGCAUAAGCAGCAUACGCAGCUGCCUGAAAGCCUGUGGAAGAAAU------GAAA ...((-......((....))...((.((.(((...((((...)))).))).)).))))..(((.((((.((.(((((.......))))).))...)))).))).......------.... ( -32.50) >DroEre_CAF1 6927 105 + 1 GAGCCGCACUUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAGGCAGCGUAGGCAGCUGCCGGAAAACCUA---------------UGGAA ..((((((((....))).(((((((((.((((.......)))).))))...))))).......)))))...((((((.......))))))((....))..---------------..... ( -40.80) >DroWil_CAF1 10458 120 + 1 GGAUAGAACUUACCAGUUGGAUAGGGUAGACCAAAGCUAGGAUAGCCGGAAUAUCCGCGACCAGCGGCAUAGGCAGCAUAAGCAGCUGCUGGAAAGCCUAAAAGAGAAUGAUGAAAAGAA ........(((....(((((...(((((..((...((((...)))).))..)))))....)))))(((...((((((.......)))))).....))).....))).............. ( -31.50) >DroAna_CAF1 6145 112 + 1 GCGUCGGACUUACCAGUUGGAUAGGGCAGACCGAAGCUGGGAUAGCCCGAGUAGCCACGACCAGCGGCAUAAGCAGCAUAGGCAGCUGCCGGAAAACCUGCACA--AAAU------AAAA ((.((((.((..((((((.....((.....))..))))))...)).))))...(((.(.....).)))....)).(((..(((....)))((....)))))...--....------.... ( -33.70) >consensus GAACCGAACUUACCAGUUGGAUAGGGCAGUCCAAAGCUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAAGCAGCAUAGGCAGCUGCCGGAAAACCUAAAAGA_AAAG______AGAA ............((....)).((((((.((((.......)))).))(((.....((((.....)))).....(((((.......))))))))....)))).................... (-22.02 = -22.22 + 0.19)


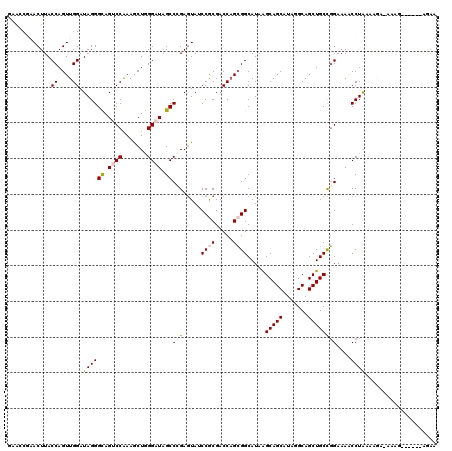
| Location | 17,039,438 – 17,039,555 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17039438 117 - 23771897 UUCUAUA---CUUUUUCGAUUAGGUUUUCCGGCAGCUGCCUAUGCUGCUUAUGCCGCUGGUCGCGGAUACUCGGGCUAUCCCAGUUUUGGACUGCCCUAUCCAACUGGUAAGUGCGGCUC .......---............(((.....((((((.......))))))...)))...(((((((((((...((((..(((.......)))..))))))))).(((....))))))))). ( -38.40) >DroVir_CAF1 1612 114 - 1 U-GUU-----UUCUUUCUCGCAGGCUUUCCGGCAGCUGCGUAUGCUGCCUAUGCCGCAGGUCGCGGCUACUCCGGCUAUCCGAGCUUCGGACUACCCUAUCCAACUGGUAAGUUUGUUUA .-...-----.........((((((((.((((((((.......))))))...(((((.....)))))......((.((((((.....)))).)).)).........)).))))))))... ( -38.40) >DroGri_CAF1 7097 113 - 1 UUUC------AUUUCUUCCACAGGCUUUCAGGCAGCUGCGUAUGCUGCUUAUGCCUCUGGUCGCGGUUACUCCGGCUAUCCUAGCUUUGGACUACCCUAUCCAACUGGUAAGUU-GCUCA ....------.......(((.((((....(((((((.......)))))))..)))).)))....(((...((((((((...)))))..)))..))).....(((((....))))-).... ( -32.50) >DroEre_CAF1 6927 105 - 1 UUCCA---------------UAGGUUUUCCGGCAGCUGCCUACGCUGCCUAUGCCGCUGGUCGCGGAUACUCGGGCUAUCCCAGUUUUGGACUGCCCUAUCCAACUGGUAAGUGCGGCUC .....---------------..(((.....((((((.......))))))...)))...(((((((((((...((((..(((.......)))..))))))))).(((....))))))))). ( -39.70) >DroWil_CAF1 10458 120 - 1 UUCUUUUCAUCAUUCUCUUUUAGGCUUUCCAGCAGCUGCUUAUGCUGCCUAUGCCGCUGGUCGCGGAUAUUCCGGCUAUCCUAGCUUUGGUCUACCCUAUCCAACUGGUAAGUUCUAUCC ....................(((((((.((((.(((((.....((((...(((((((.....)))).)))..)))).....)))))((((..........)))))))).))).))))... ( -27.40) >DroAna_CAF1 6145 112 - 1 UUUU------AUUU--UGUGCAGGUUUUCCGGCAGCUGCCUAUGCUGCUUAUGCCGCUGGUCGUGGCUACUCGGGCUAUCCCAGCUUCGGUCUGCCCUAUCCAACUGGUAAGUCCGACGC ....------....--...((.(((.....((((((.......))))))...)))(((((..((((((.....)))))).))))).((((.((..((.........))..)).)))).)) ( -37.10) >consensus UUCU______AUUU_UCGCGCAGGCUUUCCGGCAGCUGCCUAUGCUGCCUAUGCCGCUGGUCGCGGAUACUCCGGCUAUCCCAGCUUUGGACUACCCUAUCCAACUGGUAAGUUCGACUC ......................(((.....((((((.......))))))...)))(((((..((..........))....)))))..((((((..((.........))..)))))).... (-21.25 = -21.67 + 0.42)

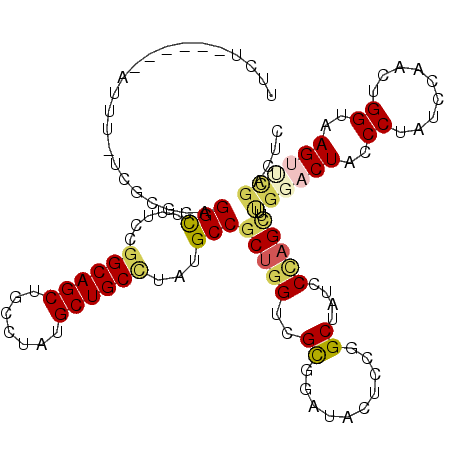
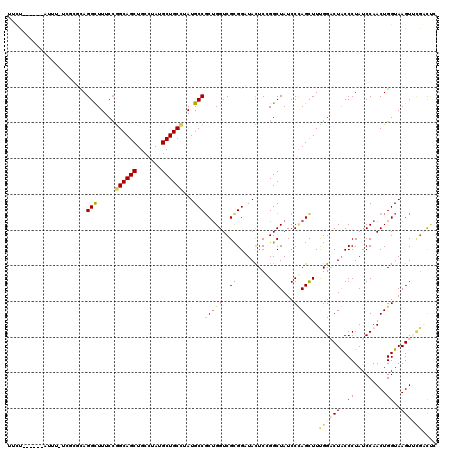
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:38 2006