
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,013,404 – 17,013,516 |
| Length | 112 |
| Max. P | 0.962371 |

| Location | 17,013,404 – 17,013,516 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.34 |
| Mean single sequence MFE | -16.93 |
| Consensus MFE | -10.08 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
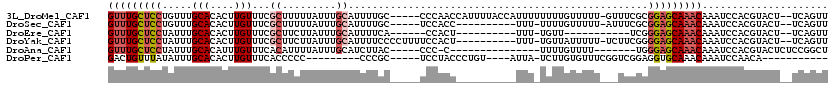
>3L_DroMel_CAF1 17013404 112 + 23771897 GUUUGCUCCUGUUUGCACACUUGUUUCGCUUUUUAUUUGCAUUUUGC-----CCCAACCAUUUUACCAUUUUUUUUGUUUUU-GUUUCGCGGAGCAAACAAAUCCACGUACU--UCAGUU (((((((((((...(((..........((.........)).......-----..............((.......))....)-))..)).))))))))).............--...... ( -16.50) >DroSec_CAF1 2351 101 + 1 GUUUGCUCCUGUUUGCACACUUGUUUCGCUUUUUAUUUGCAUUUUGC-----UCCACC----------UUU-UUUUGUUUUU-AUUUCGCGGAGCAAACAAAUCCACGUACU--UCAGUU (((((((((.((..(((....)))...)).........((.....))-----......----------...-..........-.......))))))))).............--...... ( -16.60) >DroEre_CAF1 2242 90 + 1 GUUUGCUCCUAUUUGCACACUUGUUUCGCUUCUUAUUUGCAUUUUCA------CCACU----------UUU-UGUU-----------UCGGGAGCAAACAAAUCCACGUACU--UCAGUU ((((((((((....(((....)))...((.........)).......------.....----------...-....-----------..)))))))))).............--...... ( -17.00) >DroYak_CAF1 2305 106 + 1 GUUUGCUCCUAUUUGCACACUUGUUUCGCUUCUUAUUUGCAUUUUCCCCUUUUCCACU----------UUU-UGUUAUUUUU-UCUUCGGGGAGCAAACAAAUCCACGUACU--UCAGUU (((((((((....((((.....(........).....)))).................----------...-..........-.(....)))))))))).............--...... ( -17.50) >DroAna_CAF1 1865 92 + 1 GUUUGCUCCUAUUUGCACAUUUGUUUCACAUUUUAUUUGCAUCUUAC-----CCC-C---------------UUUUGUUUU-------UGGGAGCAAACAAAUCCACGUACUCUCCGGCU (((((((((((..((((.((.((.....))....)).))))......-----...-.---------------.........-------)))))))))))..................... ( -17.90) >DroPer_CAF1 8052 90 + 1 GACUGUUUAUAUUUGCACACUUGUUUCACCCCC---------CCCGC-----UCCUACCCUGU----AUUA-UCUUGUGUUUCGGUCGGAGGUGCAAACAAAUCCAACA----------- ....................((((((((((((.---------.(((.-----...(((...(.----....-.)..)))...)))..)).)))).))))))........----------- ( -16.10) >consensus GUUUGCUCCUAUUUGCACACUUGUUUCGCUUCUUAUUUGCAUUUUGC_____UCCACC__________UUU_UUUUGUUUUU___UUCGGGGAGCAAACAAAUCCACGUACU__UCAGUU (((((((((.....(((....)))...((.........))..................................................)))))))))..................... (-10.08 = -10.97 + 0.89)


| Location | 17,013,404 – 17,013,516 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.34 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -8.65 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
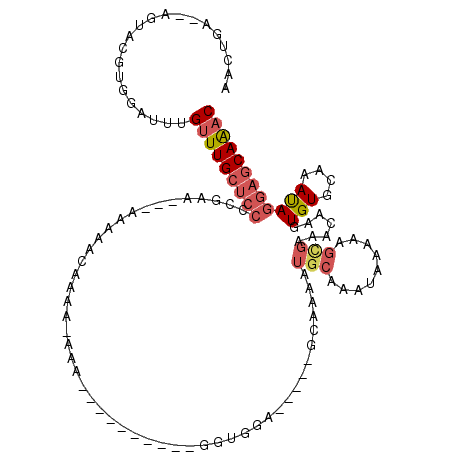
>3L_DroMel_CAF1 17013404 112 - 23771897 AACUGA--AGUACGUGGAUUUGUUUGCUCCGCGAAAC-AAAAACAAAAAAAAUGGUAAAAUGGUUGGG-----GCAAAAUGCAAAUAAAAAGCGAAACAAGUGUGCAAACAGGAGCAAAC ......--.............(((((((((((....(-((...((...............)).)))..-----((.....)).........))........(((....)))))))))))) ( -18.26) >DroSec_CAF1 2351 101 - 1 AACUGA--AGUACGUGGAUUUGUUUGCUCCGCGAAAU-AAAAACAAAA-AAA----------GGUGGA-----GCAAAAUGCAAAUAAAAAGCGAAACAAGUGUGCAAACAGGAGCAAAC ..(((.--.((((....(((((((((((((((.....-..........-...----------.)))))-----))))...)))))).....((.......))))))...)))........ ( -22.21) >DroEre_CAF1 2242 90 - 1 AACUGA--AGUACGUGGAUUUGUUUGCUCCCGA-----------AACA-AAA----------AGUGG------UGAAAAUGCAAAUAAGAAGCGAAACAAGUGUGCAAAUAGGAGCAAAC ......--.............(((((((((((.-----------.((.-...----------.))..------))....((((........((.......)).))))....))))))))) ( -16.50) >DroYak_CAF1 2305 106 - 1 AACUGA--AGUACGUGGAUUUGUUUGCUCCCCGAAGA-AAAAAUAACA-AAA----------AGUGGAAAAGGGGAAAAUGCAAAUAAGAAGCGAAACAAGUGUGCAAAUAGGAGCAAAC .(((..--....(((...((((((((((((((.....-..........-...----------.........)))))....)))))))))..))).....))).(((........)))... ( -20.33) >DroAna_CAF1 1865 92 - 1 AGCCGGAGAGUACGUGGAUUUGUUUGCUCCCA-------AAAACAAAA---------------G-GGG-----GUAAGAUGCAAAUAAAAUGUGAAACAAAUGUGCAAAUAGGAGCAAAC .((((.....(((((..((((((((((((((.-------.........---------------)-)))-----))))...))))))...))))).......)).)).............. ( -20.70) >DroPer_CAF1 8052 90 - 1 -----------UGUUGGAUUUGUUUGCACCUCCGACCGAAACACAAGA-UAAU----ACAGGGUAGGA-----GCGGG---------GGGGGUGAAACAAGUGUGCAAAUAUAAACAGUC -----------(((...((((((((.(((((((..(((...(((....-....----.....)).)..-----.))).---------)))))))))))))))..)))............. ( -24.22) >consensus AACUGA__AGUACGUGGAUUUGUUUGCUCCCCGAA___AAAAACAAAA_AAA__________GGUGGA_____GCAAAAUGCAAAUAAAAAGCGAAACAAGUGUGCAAAUAGGAGCAAAC .....................(((((((((.................................................(((.........))).......(((....)))))))))))) ( -8.65 = -9.52 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:29 2006