| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,880,325 – 1,880,423 |
| Length | 98 |
| Max. P | 0.530820 |

| Location | 1,880,325 – 1,880,423 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.01 |
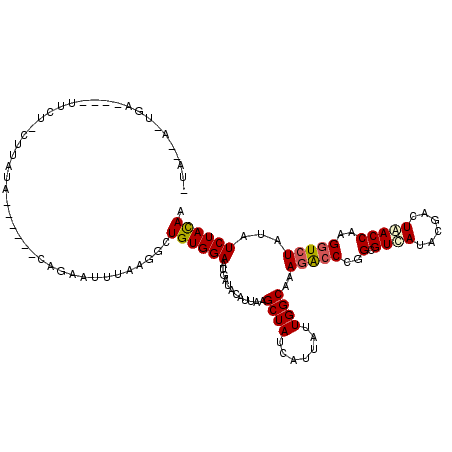
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1880325 98 - 23771897 -U--AA-------------AAACUU------UAGAACUUAAGGCUGUGGAAUGAUACACUGAGCUAUCAUUAUUGGCAAAGGCCUGGCGUCAUCCGACUGACCAAGCUAUAUAUCUACAA -.--..-------------......------((((.......((((...(((((((........)))))))..))))...(((.(((((((....))).).))).))).....))))... ( -19.50) >DroVir_CAF1 3274 109 - 1 -CA----UGA----UUCUUCGUAUAUUCG--CAGAAUUUAAGGCUGUGGAAUGAUACAUUAAGCUAUCAUUAUUGGCAAAGACCUGGCGUCAUACGUCUAACCAAGGUCUAUAUCUAUAA -..----.((----((((.((......))--.))))))....((((...(((((((........)))))))..))))..((((((((((.....))))......)))))).......... ( -23.90) >DroPse_CAF1 2778 111 - 1 -UAACA-UUCUCGCUUCU-CUUCUU------UAGAACCUAAGGCUGUGGAAUGAUACAUUGAGCUAUCAUUAUUGGCAAAGACCCGGCGUCAUACGGCUGACCAAGGUCUACAUCUAUAA -...((-(((.(((....-...(((------(((...))))))..)))))))).........((((.......))))..(((((..(.((((......)))))..))))).......... ( -23.60) >DroGri_CAF1 3285 109 - 1 -CAAAAAUGC----UACUCCAAAUA------CAGAAUUUAAGGCUGUGGAAUGACACAUUAAGCUAUCAUUAUUGGCAAAGGCCCGGCGUUAUACGCCUAACCAAGGUCUAUAUCUACAA -.........----...........------.............((((((..(((.......((((.......))))...((...((((.....))))...))...)))....)))))). ( -21.10) >DroWil_CAF1 5876 111 - 1 AUAAUA-UGAUCAUUUCG-AUU-GA------CAGAAUUUAAGGCUGUGGAAUGAUACACUAAGCUAUCAUUAUUGGCAAAGACCCGGAGUCAUACGACUAACCAAGGUCUAUAUCUAUAA ......-((((((((((.-...-.(------(((.........)))))))))))).))....((((.......))))..(((((.((((((....))))..))..))))).......... ( -22.60) >DroMoj_CAF1 4252 109 - 1 -UA----AGA-----CCUUCUUA-AUUCGAACAGAAUUUAAGGCUGUGGAAUGAUACAUUAAGCUAUCAUUAUUGGCAGAGACCCGGCGUCAUUCGGCUGACCAAGGUCUACAUCUACAA -..----((.-----((((...(-((((.....))))).))))))(((((.((.........((((.......))))..(((((..(.((((......)))))..))))).))))))).. ( -27.90) >consensus _UA__A_UGA____UUCU_CUUAUA______CAGAAUUUAAGGCUGUGGAAUGAUACAUUAAGCUAUCAUUAUUGGCAAAGACCCGGCGUCAUACGACUAACCAAGGUCUAUAUCUACAA ............................................((((((............((((.......))))..(((((..(.((((......)))))..)))))...)))))). (-15.99 = -16.02 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:18 2006