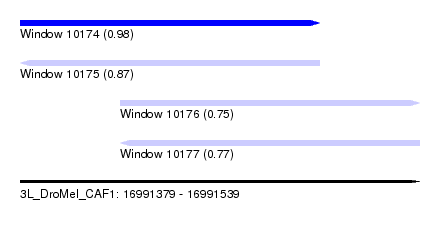
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,991,379 – 16,991,539 |
| Length | 160 |
| Max. P | 0.982637 |

| Location | 16,991,379 – 16,991,499 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -51.10 |
| Consensus MFE | -49.78 |
| Energy contribution | -48.90 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
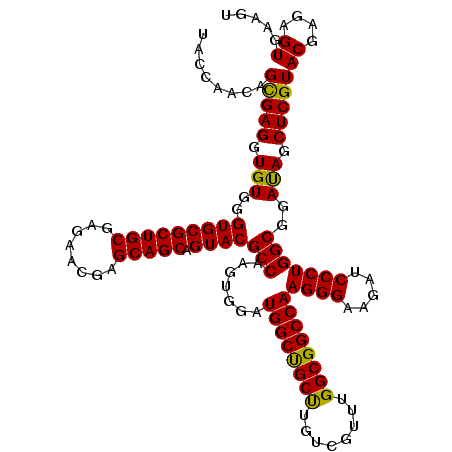
>3L_DroMel_CAF1 16991379 120 + 23771897 UAUCAACAGUGAGGUGUGGGUGCGCUGCGAGAACGAGCAGCAGUACGCCAAGUGGAUGGCUGCUUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGAUAGCUCGUACGAGAGUGAAGU ..........(((.(((..(((((((((........))))).))))(((.......((((((((........))))))))((((.....)))))))..))).))).(((....))).... ( -46.60) >DroSim_CAF1 2941 120 + 1 UACCAACAGCGAGGUGUGGGUGCGCUGCGAGAACGAGCAGCAGUACGCCAAGUGGAUGGCUGCCUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGACAGCUCGUACGAGAGUGAAGU ........(((((.(((..(((((((((........))))).))))(((.......((((((((........))))))))((((.....)))))))..))).)))))((....))..... ( -53.50) >DroYak_CAF1 3134 120 + 1 AACCAACAGCGAGGUGUGGGUGCGCUGCGAGAACGAGCAGCAGUACGCCAAGUGGAUGGCCGCUUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGAUAGCUCGUACGAGGGUGAAGU .(((....(((((.(((..(((((((((........))))).))))(((.......((((((((........))))))))((((.....)))))))..))).))))).....)))..... ( -53.20) >consensus UACCAACAGCGAGGUGUGGGUGCGCUGCGAGAACGAGCAGCAGUACGCCAAGUGGAUGGCUGCUUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGAUAGCUCGUACGAGAGUGAAGU ........(((((.(((..(((((((((........))))).))))(((.......((((((((........))))))))((((.....)))))))..))).)))))((....))..... (-49.78 = -48.90 + -0.88)

| Location | 16,991,379 – 16,991,499 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -35.89 |
| Energy contribution | -36.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
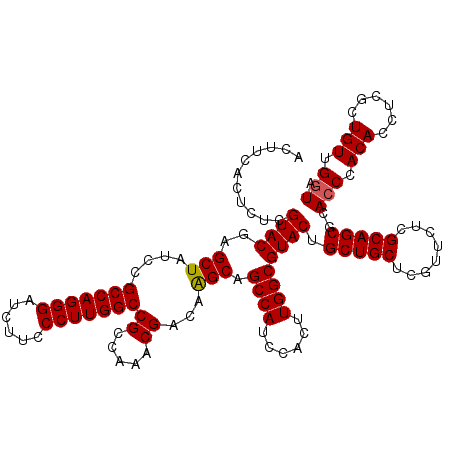
>3L_DroMel_CAF1 16991379 120 - 23771897 ACUUCACUCUCGUACGAGCUAUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAAGCAGCCAUCCACUUGGCGUACUGCUGCUCGUUCUCGCAGCGCACCCACACCUCACUGUUGAUA ...(((.....((((..(((....(((((((......)))))))((.....))...))).((((......)))))))).(((((........)))))..................))).. ( -34.50) >DroSim_CAF1 2941 120 - 1 ACUUCACUCUCGUACGAGCUGUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAGGCAGCCAUCCACUUGGCGUACUGCUGCUCGUUCUCGCAGCGCACCCACACCUCGCUGUUGGUA .............((((((.((.((((((((.....))).((((.(((........))).))))......))))).))....)))))).((.((((((...........)))))).)).. ( -41.90) >DroYak_CAF1 3134 120 - 1 ACUUCACCCUCGUACGAGCUAUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAAGCGGCCAUCCACUUGGCGUACUGCUGCUCGUUCUCGCAGCGCACCCACACCUCGCUGUUGGUU .....(((...(.((((((....((((((((.....))).(((((((..........)))))))......))))).......)))))).)..((((((...........)))))).))). ( -41.70) >consensus ACUUCACUCUCGUACGAGCUAUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAAGCAGCCAUCCACUUGGCGUACUGCUGCUCGUUCUCGCAGCGCACCCACACCUCGCUGUUGGUA ...........((((..(((....(((((((......)))))))((.....))...))).((((......)))))))).(((((........)))))..(((.(((......))).))). (-35.89 = -36.00 + 0.11)

| Location | 16,991,419 – 16,991,539 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -48.10 |
| Consensus MFE | -46.56 |
| Energy contribution | -45.90 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16991419 120 + 23771897 CAGUACGCCAAGUGGAUGGCUGCUUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGAUAGCUCGUACGAGAGUGAAGUGGACAGCAUACUCUCGCUACUGCAGAUGCAGCGACCCGCA (((.(((.((((..(....)..)))).))))))(((((((.(((.....)))))))((...((..(((((((((((..((.....)).)))))))).)))(((....)))))..))))). ( -45.30) >DroSim_CAF1 2981 120 + 1 CAGUACGCCAAGUGGAUGGCUGCCUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGACAGCUCGUACGAGAGUGAAGUGGACAGCAUACUCUCGCUACUGCAGAUGCAGCGACCUGCA ..((.((((.......((((((((........))))))))((((.....)))))))).)).((..(((((((((((..((.....)).)))))))).))).))...(((((....))))) ( -50.10) >DroYak_CAF1 3174 120 + 1 CAGUACGCCAAGUGGAUGGCCGCUUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGAUAGCUCGUACGAGGGUGAAGUGGAUAGCAUACUCUCGUUACUGCAGAUGCAGCGACCAGCA .....((((.......((((((((........))))))))((((.....))))))))....(((.(((((((((((..((.....)).)))))))))..((((....))))..)).))). ( -48.90) >consensus CAGUACGCCAAGUGGAUGGCUGCUUGUCGUUUGGCGGCCAAGGGAAGAUCCCUGGCGGAUAGCUCGUACGAGAGUGAAGUGGACAGCAUACUCUCGCUACUGCAGAUGCAGCGACCAGCA .....((((.......((((((((........))))))))((((.....))))))))....(((((..((((((((..((.....)).))))))))...((((....)))))))...)). (-46.56 = -45.90 + -0.66)

| Location | 16,991,419 – 16,991,539 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -44.56 |
| Consensus MFE | -40.65 |
| Energy contribution | -40.54 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16991419 120 - 23771897 UGCGGGUCGCUGCAUCUGCAGUAGCGAGAGUAUGCUGUCCACUUCACUCUCGUACGAGCUAUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAAGCAGCCAUCCACUUGGCGUACUG (((..((((((((....))))).((((((((..............))))))))...........(((((((......)))))))........)))..)))((((......))))...... ( -43.34) >DroSim_CAF1 2981 120 - 1 UGCAGGUCGCUGCAUCUGCAGUAGCGAGAGUAUGCUGUCCACUUCACUCUCGUACGAGCUGUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAGGCAGCCAUCCACUUGGCGUACUG .((((.(((((((....))))).((((((((..............))))))))..)).)))).((((((((.....))).((((.(((........))).))))......)))))..... ( -47.94) >DroYak_CAF1 3174 120 - 1 UGCUGGUCGCUGCAUCUGCAGUAACGAGAGUAUGCUAUCCACUUCACCCUCGUACGAGCUAUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAAGCGGCCAUCCACUUGGCGUACUG ..((.((..((((....))))..)).))(((((((((.........((((.((..((....)).)).)))).........(((((((..........)))))))......))))))))). ( -42.40) >consensus UGCAGGUCGCUGCAUCUGCAGUAGCGAGAGUAUGCUGUCCACUUCACUCUCGUACGAGCUAUCCGCCAGGGAUCUUCCCUUGGCCGCCAAACGACAAGCAGCCAUCCACUUGGCGUACUG (((..((((((((....))))).((((((((..............))))))))...........(((((((......)))))))........)))..)))((((......))))...... (-40.65 = -40.54 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:16 2006