
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,990,278 – 16,990,438 |
| Length | 160 |
| Max. P | 0.981760 |

| Location | 16,990,278 – 16,990,398 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -50.50 |
| Consensus MFE | -47.69 |
| Energy contribution | -48.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16990278 120 + 23771897 UAUCCGCUAUCCGGAGGAGUUGUCCCUGUGCAAACCACUGGAAGCGGAGCACCUGAAGCGUAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUCGCUGAACCAGAUGGCACCAC ..((((((.(((((.((..((((......)))).)).)))))))))))(((((((..(((......))).)))))))(((.......))).((((((....(((...))).))))))... ( -49.40) >DroSim_CAF1 1840 120 + 1 UAUCCGCUAUCCGGAGGAGUUGUCCCUGUGCAAACCUCUGGAAGCGGAGCACCUGAAGCGUAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUUGCUGAGCCAGAUGGCACCAC ..((((((.((((((((..((((......)))).))))))))))))))(((((((..(((......))).)))))))(((.......))).((((((.(((......))).))))))... ( -55.00) >DroYak_CAF1 2033 120 + 1 UAUCCGCUACCCGGAGGAGUUGUCCCUGUGCAAACCUCUGGAAGCCGAGCACCUGAAGCGCAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUCGCUGAGCCCGAUGGCACCAC ....((((..(((((((..((((......)))).)))))))..((.(.(((((((..(((......))).))))))))))......)))).((((((.(((.......)))))))))... ( -47.10) >consensus UAUCCGCUAUCCGGAGGAGUUGUCCCUGUGCAAACCUCUGGAAGCGGAGCACCUGAAGCGUAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUCGCUGAGCCAGAUGGCACCAC ..((((((.((((((((..((((......)))).))))))))))))))(((((((..(((......))).)))))))(((.......))).((((((.((.........))))))))... (-47.69 = -48.47 + 0.78)



| Location | 16,990,278 – 16,990,398 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -50.87 |
| Consensus MFE | -47.57 |
| Energy contribution | -47.80 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16990278 120 - 23771897 GUGGUGCCAUCUGGUUCAGCGAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUACGCUUCAGGUGCUCCGCUUCCAGUGGUUUGCACAGGGACAACUCCUCCGGAUAGCGGAUA ..(((((((..((......))..)))))))(((.......)))(((((((((((......))).))))))))(((((((((.(((.....))).((((....))))..))).)))))).. ( -50.20) >DroSim_CAF1 1840 120 - 1 GUGGUGCCAUCUGGCUCAGCAAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUACGCUUCAGGUGCUCCGCUUCCAGAGGUUUGCACAGGGACAACUCCUCCGGAUAGCGGAUA ..(((((((..((......))..)))))))(((.......)))(((((((((((......))).))))))))(((((((((.((((.(((........)))..)))).))).)))))).. ( -53.00) >DroYak_CAF1 2033 120 - 1 GUGGUGCCAUCGGGCUCAGCGAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUGCGCUUCAGGUGCUCGGCUUCCAGAGGUUUGCACAGGGACAACUCCUCCGGGUAGCGGAUA ((((((((((((.......))).))))))..(((((((..((((((((((((((......))).)))))))))..))...)))))))...)))..(((....)))((((.....)))).. ( -49.40) >consensus GUGGUGCCAUCUGGCUCAGCGAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUACGCUUCAGGUGCUCCGCUUCCAGAGGUUUGCACAGGGACAACUCCUCCGGAUAGCGGAUA ..(((((((....((...))...)))))))(((.......)))(((((((((((......))).))))))))(((((((((.((((.(((........)))..)))).))).)))))).. (-47.57 = -47.80 + 0.23)



| Location | 16,990,318 – 16,990,438 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -40.79 |
| Energy contribution | -41.57 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
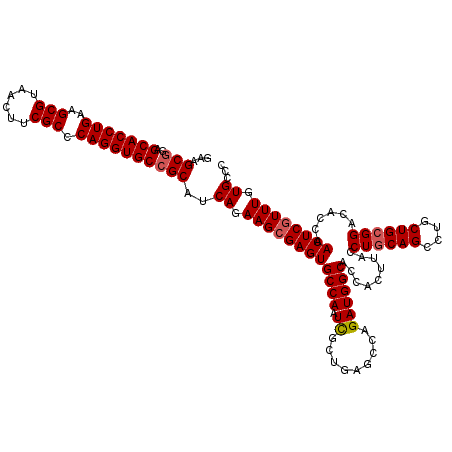
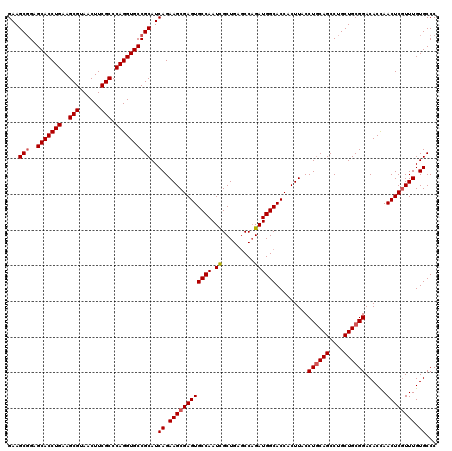
>3L_DroMel_CAF1 16990318 120 + 23771897 GAAGCGGAGCACCUGAAGCGUAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUCGCUGAACCAGAUGGCACCACUUACCUGCAGCCUGCUGCGGACACCAACUCAUUUGUGCCC ((.(((..(((((((..(((......))).)))))))))).))..(((.(.((((((....(((...))).))))))).)))..((((((....)))))).................... ( -42.30) >DroSim_CAF1 1880 120 + 1 GAAGCGGAGCACCUGAAGCGUAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUUGCUGAGCCAGAUGGCACCACUUACCUGCAGCCUGCUGCGGACACUAACUCGUUUGUGCCC ...(((..(((((((..(((......))).))))))))))..((.((((((((((((.(((......))).)))).........((((((....))))))......)))))))).))... ( -47.80) >DroYak_CAF1 2073 120 + 1 GAAGCCGAGCACCUGAAGCGCAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUCGCUGAGCCCGAUGGCACCACUUACCUACAGCCUGCUGCGGACACCAACUCGUUUGUGCCC ...(((((((....((((.....)))).....(((((((((.((((((.(.((((((.(((.......)))))))))).))).........))).))))).)))).....))))).)).. ( -40.70) >consensus GAAGCGGAGCACCUGAAGCGUAACUUCGCCCAGGUGCCGCAUCAGAAGCGAGUGCCAAUCGCUGAGCCAGAUGGCACCACUUACCUGCAGCCUGCUGCGGACACCAACUCGUUUGUGCCC ...(((..(((((((..(((......))).))))))))))..((.((((((((((((.((.........)))))).........((((((....))))))......)))))))).))... (-40.79 = -41.57 + 0.78)

| Location | 16,990,318 – 16,990,438 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -52.37 |
| Consensus MFE | -50.41 |
| Energy contribution | -49.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
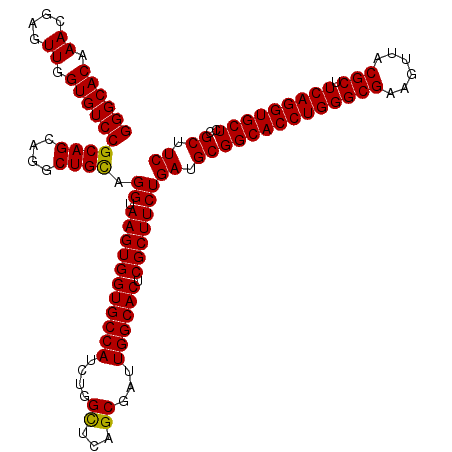
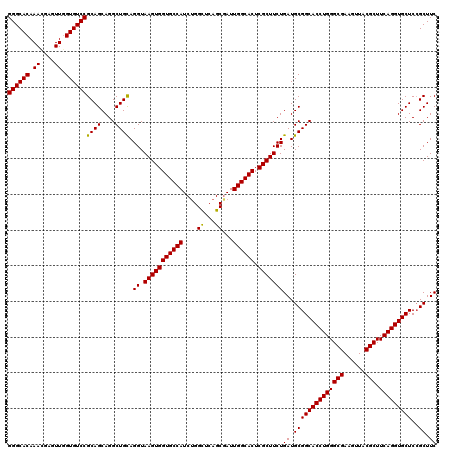
>3L_DroMel_CAF1 16990318 120 - 23771897 GGGCACAAAUGAGUUGGUGUCCGCAGCAGGCUGCAGGUAAGUGGUGCCAUCUGGUUCAGCGAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUACGCUUCAGGUGCUCCGCUUC ((((((..........))))))((((....)))).((.(((((((((((..((......))..)))))).)))))))((.((((((((((((((......))).))))))))..))).)) ( -52.00) >DroSim_CAF1 1880 120 - 1 GGGCACAAACGAGUUAGUGUCCGCAGCAGGCUGCAGGUAAGUGGUGCCAUCUGGCUCAGCAAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUACGCUUCAGGUGCUCCGCUUC ((((((.((....)).))))))((((....)))).((.(((((((((((..((......))..)))))).)))))))((.((((((((((((((......))).))))))))..))).)) ( -53.50) >DroYak_CAF1 2073 120 - 1 GGGCACAAACGAGUUGGUGUCCGCAGCAGGCUGUAGGUAAGUGGUGCCAUCGGGCUCAGCGAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUGCGCUUCAGGUGCUCGGCUUC ((((((.((....)).)))))).....((((((..((.((((((((((((((.......))).)))))).))))))).....((((((((((((......))).))))))))))))))). ( -51.60) >consensus GGGCACAAACGAGUUGGUGUCCGCAGCAGGCUGCAGGUAAGUGGUGCCAUCUGGCUCAGCGAUUGGCACUCGCUUCUGAUGCGGCACCUGGGCGAAGUUACGCUUCAGGUGCUCCGCUUC ((((((.((....)).))))))((((....)))).((.(((((((((((....((...))...)))))).)))))))((.((((((((((((((......))).)))))))))..)).)) (-50.41 = -49.97 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:12 2006