| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,979,094 – 16,979,208 |
| Length | 114 |
| Max. P | 0.821385 |

| Location | 16,979,094 – 16,979,208 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -27.01 |
| Energy contribution | -26.74 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
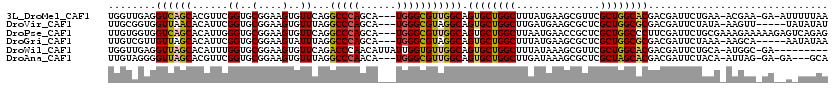
>3L_DroMel_CAF1 16979094 114 + 23771897 UGGUUGAGGUCAGCACGUUCGGUGCGGAAGUGUUCAGGCCCAGCA---UGGGCGUUGGCAGUGCUGGCUUUAUGAAGCGUUCGCUGGCACGACGAUUCUGAA-ACGAA-GA-AUUUUUAA ..((((..((((((((((((((..(....)..)).(((((.((((---(..((....)).))))))))))...).)))))..)))))).))))((((((...-....)-))-)))..... ( -38.70) >DroVir_CAF1 426 111 + 1 UUGCGGUGGUUAACACAUUCGGUGCGGAAGUGUUUAGGCCCAGCA---UGGGCGUAGGCAGUGCUGGCUUGAUGAAGCGCUCGCUGGCGCGACGAUUCUAUA-AAGUU-----UAUAUAU ((((((((.....)))..(((((((((.(.((((((.((((....---.)))).)))))).).)))((((....))))...)))))))))))..........-.....-----....... ( -34.90) >DroPse_CAF1 571 117 + 1 UUGUGGUGGUCAGCACAUUGGGUGCGGAAGUGUUCAGGCCCAGCA---UGGGCGUUGGCAGUGCUGGCUUAAUGAACCGCUCGCUGGCCCUUCGAUUCUGCGAAAGAAAAAGAGUCAGAG ..((((((((((((((.(((((..(....)..)))))((((....---.)))).......)))))))))......))))).........((..((((((.(....)....))))))..)) ( -46.30) >DroGri_CAF1 409 111 + 1 UUGUCGUUGUUAGCACAUUCGGUGCGGAAGUAUUUAGGCCCAGCA---UGGGCGUAGGCAGUGCUGGCUUUAUGAAGCGCUCGCUGGCGCGACGAUUCUAAA-AAGCA-----AAUAUAA ..((((((((((((((.....((((....))))(((.((((....---.)))).)))...))))))))........(((((....)))))))))))......-.....-----....... ( -39.40) >DroWil_CAF1 426 109 + 1 UGGUUGAGGUUAGCACAUUUGGUGCGGAAGUGUUCAGACCCAACAUUAUUGGUGUUGGCAGUGCUGGCUUUAUAAAGCGUUCGCUGGCACGACGAUUCUGCA-AUGGC-GA--------- ....((((((((((((.(((((..(....)..).)))).(((((((.....)))))))..))))))))))))........(((((.(((.((....))))).-..)))-))--------- ( -37.70) >DroAna_CAF1 399 111 + 1 UUGUAGGGGUUAGCACGUUCGGUGCGGAAGUGUUUAGGCCCAACA---UGGGCGUUGGCAGUGCUGGCUUGAUAAAGCGCUCGCUAGCACGACGAUUCUACA-AUUAG-GA-GA---GCA ((((((((((((((......((..(....)..))...((((....---.)))))))))).((((((((..(........)..)))))))).....)))))))-)....-..-..---... ( -37.70) >consensus UUGUGGUGGUUAGCACAUUCGGUGCGGAAGUGUUCAGGCCCAGCA___UGGGCGUUGGCAGUGCUGGCUUUAUGAAGCGCUCGCUGGCACGACGAUUCUACA_AAGAA_GA__A_UAUAA ........((((((......((..(....)..))...(((((......))))))))))).((((((((..............)))))))).............................. (-27.01 = -26.74 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:04 2006