
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,962,828 – 16,962,918 |
| Length | 90 |
| Max. P | 0.809215 |

| Location | 16,962,828 – 16,962,918 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -15.55 |
| Energy contribution | -17.44 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
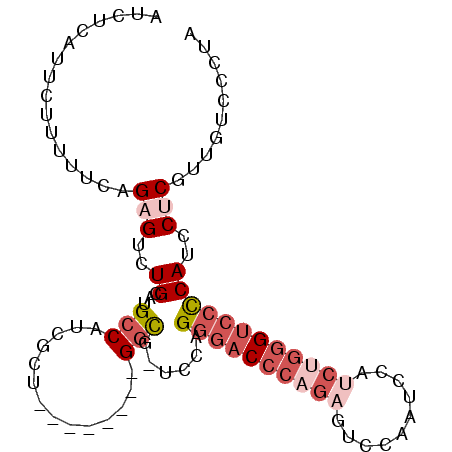
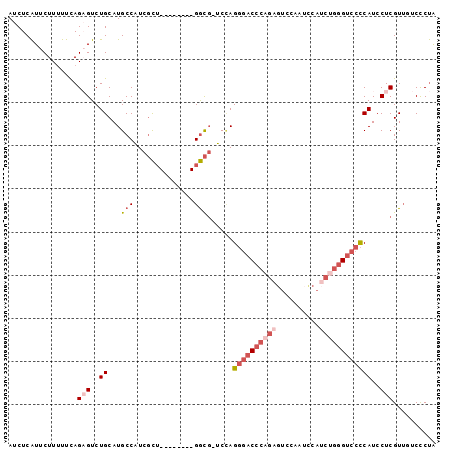
>3L_DroMel_CAF1 16962828 90 + 23771897 AUCUCAUUCUUUUUCAGAGUCUGCAUGCCAUCGCU--------GGCG-UCCAGGGACCCAGAGUCCAAACCAUCUGGGUCCCCAUCCUCGUUGUCCCUA ................(((..((.((((((....)--------))))-).))((((((((((..........))))))))))....))).......... ( -30.40) >DroSec_CAF1 1894 90 + 1 AUCUCAUUCUUUUUCAGAGUCUGCAUGCCAUCGCU--------GGCG-UCUAGGGACCCAGAGUCCAAUCCAUCUGGGUCCCCAUCCUCGUUGUCCCUA ................(((..((.((((((....)--------))))-)...((((((((((..........))))))))))))..))).......... ( -30.20) >DroSim_CAF1 1881 90 + 1 AUCUCAUUCUUUUUCAGAGUCUGCAUGCCAUCGCU--------GGCG-UCCAGGGACCCAGAGUCCAAUCCAUCUGGGUCCCCAUCCUCGUUGUCCCUA ................(((..((.((((((....)--------))))-).))((((((((((..........))))))))))....))).......... ( -30.40) >DroEre_CAF1 1891 90 + 1 AUCUCAUUCUUCUUCAGAGUCUGCAUGCCAUCGCU--------GGCG-UCCAGGGACCCUGAGUCCAAUUCAUCUGGGUCCCCAUCCUCGUUCUCCCUA ................(((..((.((((((....)--------))))-).))((((((((((((...)))))...)))))))....))).......... ( -25.80) >DroYak_CAF1 1882 90 + 1 AUCUCAUUCUUUUUUAGUGUCUGCAUGCCAUCGCU--------GGCG-UCCAGGGACCCUAGGUCCAAUCCAGCUGGGUCCCCAUCCUCGUUCUCCCUA .....................((.((((((....)--------))))-).))(((((((..((......))....)))))))................. ( -23.60) >DroPer_CAF1 1939 87 + 1 AUCUCGUUCUUUUUCAGCGUCUGCAUGCCGUCCUGAUCUGCCUGAUCUUCAAG--CCCCAG------AUCCCUCUC-GUCCUCAUCCUCGU---CUCCG ..................(((((...((......((((.....)))).....)--)..)))------)).......-..............---..... ( -8.90) >consensus AUCUCAUUCUUUUUCAGAGUCUGCAUGCCAUCGCU________GGCG_UCCAGGGACCCAGAGUCCAAUCCAUCUGGGUCCCCAUCCUCGUUGUCCCUA ................(((..((...(((..............)))......((((((((((..........))))))))))))..))).......... (-15.55 = -17.44 + 1.89)

| Location | 16,962,828 – 16,962,918 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -16.98 |
| Energy contribution | -18.87 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
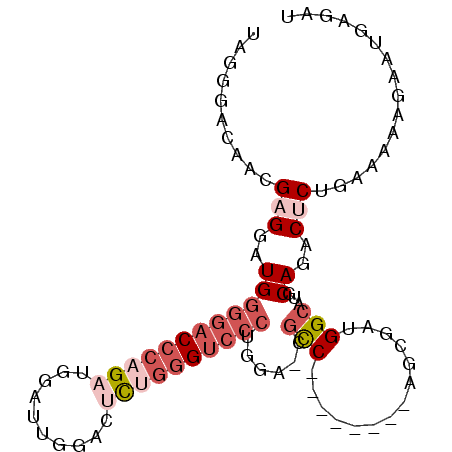
>3L_DroMel_CAF1 16962828 90 - 23771897 UAGGGACAACGAGGAUGGGGACCCAGAUGGUUUGGACUCUGGGUCCCUGGA-CGCC--------AGCGAUGGCAUGCAGACUCUGAAAAAGAAUGAGAU (((((.......(..((((((((((((.(.......)))))))))))))..-)(((--------(....)))).......))))).............. ( -34.30) >DroSec_CAF1 1894 90 - 1 UAGGGACAACGAGGAUGGGGACCCAGAUGGAUUGGACUCUGGGUCCCUAGA-CGCC--------AGCGAUGGCAUGCAGACUCUGAAAAAGAAUGAGAU (((((.......(..((((((((((((.(.......)))))))))))))..-)(((--------(....)))).......))))).............. ( -35.60) >DroSim_CAF1 1881 90 - 1 UAGGGACAACGAGGAUGGGGACCCAGAUGGAUUGGACUCUGGGUCCCUGGA-CGCC--------AGCGAUGGCAUGCAGACUCUGAAAAAGAAUGAGAU (((((.......(..((((((((((((.(.......)))))))))))))..-)(((--------(....)))).......))))).............. ( -34.30) >DroEre_CAF1 1891 90 - 1 UAGGGAGAACGAGGAUGGGGACCCAGAUGAAUUGGACUCAGGGUCCCUGGA-CGCC--------AGCGAUGGCAUGCAGACUCUGAAGAAGAAUGAGAU ...((((.....(..(((((((((...(((.......))))))))))))..-)(((--------(....)))).......))))............... ( -28.50) >DroYak_CAF1 1882 90 - 1 UAGGGAGAACGAGGAUGGGGACCCAGCUGGAUUGGACCUAGGGUCCCUGGA-CGCC--------AGCGAUGGCAUGCAGACACUAAAAAAGAAUGAGAU (((.........(..(((((((((....((......))..)))))))))..-)(((--------(....)))).........))).............. ( -26.50) >DroPer_CAF1 1939 87 - 1 CGGAG---ACGAGGAUGAGGAC-GAGAGGGAU------CUGGGG--CUUGAAGAUCAGGCAGAUCAGGACGGCAUGCAGACGCUGAAAAAGAACGAGAU ((...---.))..........(-(.......(------((((.(--((((.....)))))...))))).((((........))))........)).... ( -20.00) >consensus UAGGGACAACGAGGAUGGGGACCCAGAUGGAUUGGACUCUGGGUCCCUGGA_CGCC________AGCGAUGGCAUGCAGACUCUGAAAAAGAAUGAGAU ..........(((..((((((((((((..........))))))))))......(((..............)))...))..)))................ (-16.98 = -18.87 + 1.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:57 2006