
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
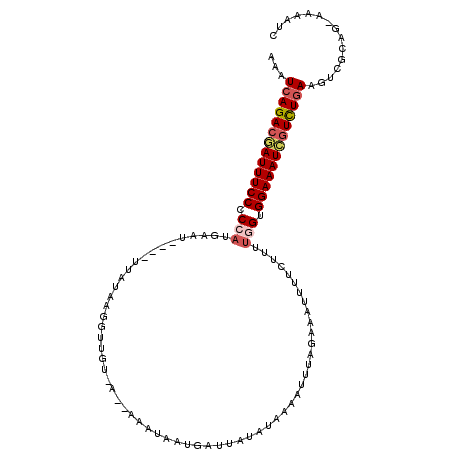
| Location | 16,955,595 – 16,955,702 |
| Length | 107 |
| Max. P | 0.999591 |

| Location | 16,955,595 – 16,955,702 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -15.67 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
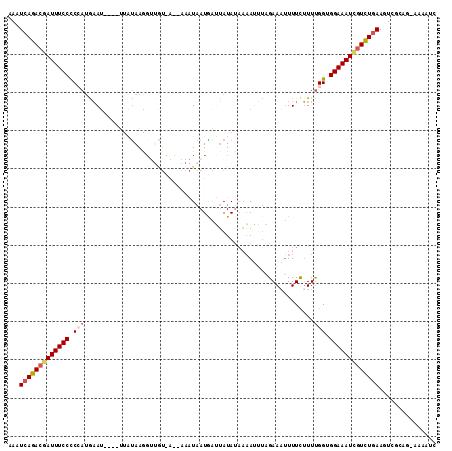
>3L_DroMel_CAF1 16955595 107 + 23771897 UAAUCAGACGAUUUCCCCGAAGUAU----UUGUAAGGUUGUUACAAAAUAAUGAUGAUAUAUAAGAUAGAAAUUUUCGUUCGGUGGAAAUCGUCUGAAGUCGUAG-AAAAUC ...((((((((((((((((((.(((----((.....((..(((........)))..)).....)))))((.....)).))))).)))))))))))))........-...... ( -32.40) >DroSec_CAF1 31000 107 + 1 UAAUCAGACGAUUUCCCCAAUGAAU----UUACAAAGUUGUCAUUAAAUAAUGGUUAUAUUAAAAUUAGAAUUUUUUUUUUGGUGGAAAUCAUUUGAAGUCGCAG-AAAAUC ...(((((.(((((((((((.((((----(((((....)))((((....))))...............)))))).....)))).))))))).)))))........-...... ( -19.30) >DroSim_CAF1 34113 107 + 1 GAAUCAGACGAUUUCCCCCAUGAAU----UUAUAAAGUUGUCAUUAAAUAAUGGUUAUAUAAAAUGUAGAAAUUUUCUUUUGGUGGAAAUCGUUUGAAGUCGCAG-AAAAUC ((.(((((((((((((.(((.....----.(((((.(((((......)))))..))))).(((((......)))))....))).)))))))))))))..))....-...... ( -25.70) >DroEre_CAF1 30773 100 + 1 AAAUGAGACUAUUUCCUCCAUAAACUUGUGCUUAAG------------UAAUACGUAUACAACAUUUCGAAAGUUUCUUUUGGAGGAAAUGGUCUGAAGUCGCAGUAAAAUC ...(.(((((((((((((((...(((((....))))------------)..................(....).......))))))))))))))).)............... ( -27.50) >DroYak_CAF1 32051 101 + 1 AAGUCAGACGAUUUCCUCCAUGAAU----AAAUAAGGCCAU-------UAAUAAGUAUAUAAAAUUCCGAAAUUUUCUUUUGGUGGAAAUGGUCUGAAGUCGCAAUCAAAUC ...((((((.((((((.(((.(((.----......((....-------..(((....)))......))......)))...))).)))))).))))))............... ( -23.12) >consensus AAAUCAGACGAUUUCCCCCAUGAAU____UUAUAAGGUUGU_A__AAAUAAUGAUUAUAUAAAAUUUAGAAAUUUUCUUUUGGUGGAAAUCGUCUGAAGUCGCAG_AAAAUC ...(((((((((((((.(((............................................................))).)))))))))))))............... (-15.67 = -16.39 + 0.72)

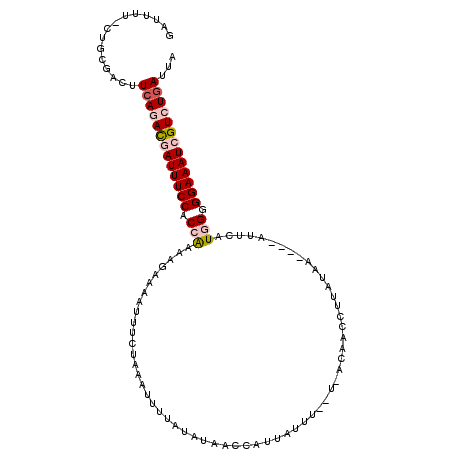
| Location | 16,955,595 – 16,955,702 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -13.53 |
| Energy contribution | -14.61 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
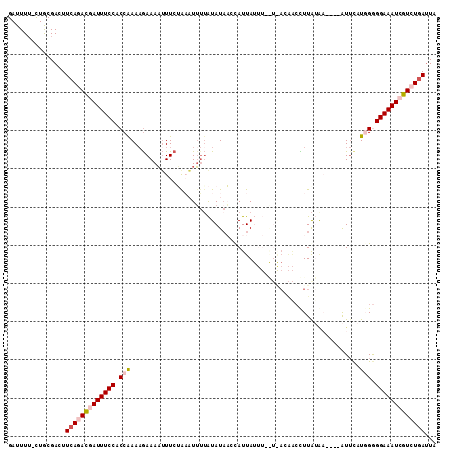
>3L_DroMel_CAF1 16955595 107 - 23771897 GAUUUU-CUACGACUUCAGACGAUUUCCACCGAACGAAAAUUUCUAUCUUAUAUAUCAUCAUUAUUUUGUAACAACCUUACAA----AUACUUCGGGGAAAUCGUCUGAUUA ......-........(((((((((((((.(((((.((.....)).....................(((((((.....))))))----)...))))))))))))))))))... ( -30.70) >DroSec_CAF1 31000 107 - 1 GAUUUU-CUGCGACUUCAAAUGAUUUCCACCAAAAAAAAAAUUCUAAUUUUAAUAUAACCAUUAUUUAAUGACAACUUUGUAA----AUUCAUUGGGGAAAUCGUCUGAUUA ......-........(((.(((((((((.((((..........................((((....))))(((....)))..----.....))))))))))))).)))... ( -18.70) >DroSim_CAF1 34113 107 - 1 GAUUUU-CUGCGACUUCAAACGAUUUCCACCAAAAGAAAAUUUCUACAUUUUAUAUAACCAUUAUUUAAUGACAACUUUAUAA----AUUCAUGGGGGAAAUCGUCUGAUUC ......-........(((.(((((((((.(((..(((.....))).((((..(((.......)))..))))............----.....))).))))))))).)))... ( -21.60) >DroEre_CAF1 30773 100 - 1 GAUUUUACUGCGACUUCAGACCAUUUCCUCCAAAAGAAACUUUCGAAAUGUUGUAUACGUAUUA------------CUUAAGCACAAGUUUAUGGAGGAAAUAGUCUCAUUU .................((((.((((((((((...............((((.....))))....------------..(((((....))))))))))))))).))))..... ( -22.20) >DroYak_CAF1 32051 101 - 1 GAUUUGAUUGCGACUUCAGACCAUUUCCACCAAAAGAAAAUUUCGGAAUUUUAUAUACUUAUUA-------AUGGCCUUAUUU----AUUCAUGGAGGAAAUCGUCUGACUU ...............((((((.((((((.(((..((.((((...((......(((....)))..-------....))..))))----.))..))).)))))).))))))... ( -20.70) >consensus GAUUUU_CUGCGACUUCAGACGAUUUCCACCAAAAGAAAAUUUCUAAAUUUUAUAUAACCAUUAUUU__U_ACAACCUUAUAA____AUUCAUGGGGGAAAUCGUCUGAUUA ...............(((((((((((((.(((............................................................))).)))))))))))))... (-13.53 = -14.61 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:54 2006