| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,895,248 – 16,895,342 |
| Length | 94 |
| Max. P | 0.770998 |

| Location | 16,895,248 – 16,895,342 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.23 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -8.71 |
| Energy contribution | -10.62 |
| Covariance contribution | 1.91 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16895248 94 + 23771897 CAUUUUCAGCAACUGAUUUCAUAGGAAAAUUGUCUUUGCGUGUUUUGCGUACUGUGUCAAUUGACUUUCAAAAU----AAUGCCAAGUAAUCAGUUAA ..........((((((((.....((...(((((...(((((.....)))))....(((....))).......))----))).))....)))))))).. ( -16.80) >DroSec_CAF1 6903 94 + 1 CAUUUUCAGCAACUGAUUUCUUAGGAACAUUUUCUUUGGGUGUUUUGCGUACUGUGUCAAUUGACUUUCAAAAU----AAUGCAAAGAAAUCAGUUGA .........(((((((((((((.(((((((((.....)))))))))((((..((.(((....)))...))....----.)))).))))))))))))). ( -26.80) >DroSim_CAF1 6619 94 + 1 CAUAUUCAGCAACUGAUUUCUUAGGAAAACUUUCUUUGUGUGUUUUGCGUACUGUAUCAAUUGACCUUCAAAAU----AAUGCUAAGUAAUCAGUUGA .........(((((((((.(((((.(...........(((((.....)))))........(((.....)))...----..).))))).))))))))). ( -18.60) >DroEre_CAF1 7177 93 + 1 CACUUUCAGGAACCGAUUUCUUGGGAACAUUUUCUCUUGGUGUUUGGGUUACUG-GCCAAUUGACUUUCAAAAU----AGUGCUAAGAAAUCAGUUAA ..........(((.(((((((((((((((((.......))))))).(((.....-)))................----....)))))))))).))).. ( -20.40) >DroYak_CAF1 7524 79 + 1 CACUUUCAGCCACCGAUUUCUUAGGAUUAUUUUG------------------UG-GCCAAAUGACUAUCGAAAUCUAAGAUGCUAAGAAAUCAGGUAA ...........(((((((((((((.(((((((((------------------((-(.(....).))).))))))....))).)))))))))).))).. ( -20.40) >consensus CAUUUUCAGCAACUGAUUUCUUAGGAAAAUUUUCUUUGGGUGUUUUGCGUACUGUGUCAAUUGACUUUCAAAAU____AAUGCUAAGAAAUCAGUUAA ..........((((((((((((((.(.............(((.......)))........(((.....))).........).)))))))))))))).. ( -8.71 = -10.62 + 1.91)


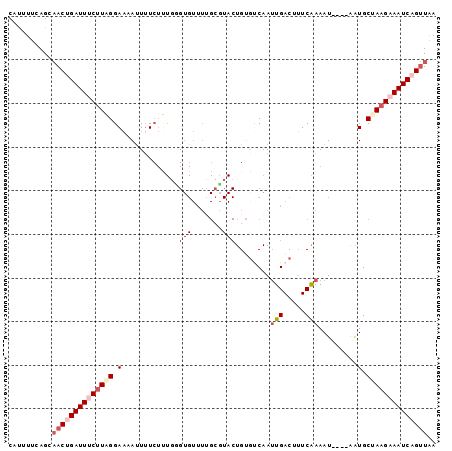
| Location | 16,895,248 – 16,895,342 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 76.23 |
| Mean single sequence MFE | -17.72 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16895248 94 - 23771897 UUAACUGAUUACUUGGCAUU----AUUUUGAAAGUCAAUUGACACAGUACGCAAAACACGCAAAGACAAUUUUCCUAUGAAAUCAGUUGCUGAAAAUG .(((((((((.(..((....----(..(((...(((....))).......((.......)).....)))..).))...).)))))))))......... ( -14.00) >DroSec_CAF1 6903 94 - 1 UCAACUGAUUUCUUUGCAUU----AUUUUGAAAGUCAAUUGACACAGUACGCAAAACACCCAAAGAAAAUGUUCCUAAGAAAUCAGUUGCUGAAAAUG .(((((((((((((.(((((----.(((((...(((....)))...((.......))...)))))..)))))....)))))))))))))......... ( -22.70) >DroSim_CAF1 6619 94 - 1 UCAACUGAUUACUUAGCAUU----AUUUUGAAGGUCAAUUGAUACAGUACGCAAAACACACAAAGAAAGUUUUCCUAAGAAAUCAGUUGCUGAAUAUG .(((((((((.(((((.(..----(((((....(((....)))...((.......))........)))))..).))))).)))))))))......... ( -16.80) >DroEre_CAF1 7177 93 - 1 UUAACUGAUUUCUUAGCACU----AUUUUGAAAGUCAAUUGGC-CAGUAACCCAAACACCAAGAGAAAAUGUUCCCAAGAAAUCGGUUCCUGAAAGUG ..((((((((((((((((..----.(((((...((...((((.-.......))))))..))))).....))))...)))))))))))).......... ( -16.50) >DroYak_CAF1 7524 79 - 1 UUACCUGAUUUCUUAGCAUCUUAGAUUUCGAUAGUCAUUUGGC-CA------------------CAAAAUAAUCCUAAGAAAUCGGUGGCUGAAAGUG .((((.((((((((((.......((((.....)))).((((..-..------------------))))......)))))))))))))).......... ( -18.60) >consensus UUAACUGAUUUCUUAGCAUU____AUUUUGAAAGUCAAUUGACACAGUACGCAAAACACCCAAAGAAAAUGUUCCUAAGAAAUCAGUUGCUGAAAAUG .(((((((((((((((.................(((....)))...((.......)).................)))))))))))))))......... (-12.70 = -13.52 + 0.82)

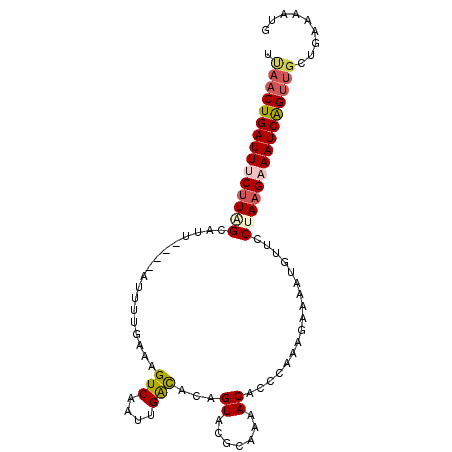
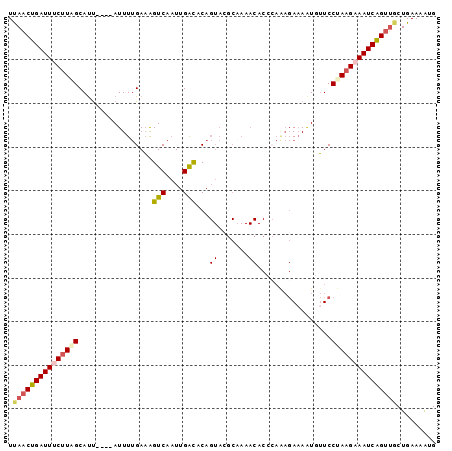
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:42 2006