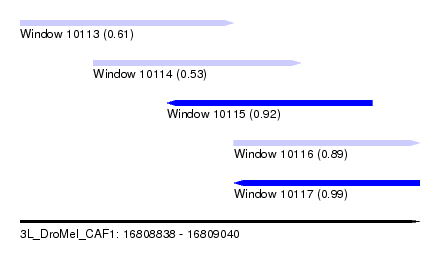
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,808,838 – 16,809,040 |
| Length | 202 |
| Max. P | 0.994744 |

| Location | 16,808,838 – 16,808,946 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.71 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.32 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16808838 108 + 23771897 UGGUGGUAGCAACUUUCCACCGUAUCCCACGGGAUCUAAUGUCGGACCGUACCCCCCGACACCCGCUGGUCCAGCUGGC---GGAUCCGGAUAUCCGGCCUACCCGAAUUU .(((((..........)))))........((((......((((((..........)))))).((((..(.....)..))---))..((((....))))....))))..... ( -39.80) >DroVir_CAF1 3297 108 + 1 UAGCGGUGGUAACUUUCCGCCAUAUCCCACUGGUGGCAACUAUAUGCCCUAUCCGGCGACU---GGUGGACCGGCAGGAAACUCCGCCGGCUAUCAGCCCUACAUGAAUUA ....(((((.......)))))......(.((((.((((......))))....)))).).((---(((((.(((((((....))..)))))))))))).............. ( -39.50) >DroGri_CAF1 3029 111 + 1 UAGCGGUGGUAACUUUCCGCCCUAUCCCACUGGCGGCACUUUCAUGCCAUAUCCAUCGACCAGUGCUGGACCGACUGGUAGUUCAGGCGGAUAUCCGCCGUACAUGAAUUA ..(((((((......((((((....((((((((.((((......))))...((....))))))))..))(((....)))......))))))...))))))).......... ( -38.80) >DroYak_CAF1 868 108 + 1 UGGUGGCGGCAACUUUCCACCGUAUCCCACAGGAUUCAACUUCGAACCGUAUCCCCCGACACCUGCUGGUCCAGCUGGC---GGAUCCGGCUAUCCGCCGUACAUGAAUUU .(((((.(....)...)))))((((.(((((((.(((......)))..((........)).)))).))).......(((---((((......)))))))))))........ ( -31.70) >DroAna_CAF1 1040 111 + 1 UGGUGGCGGCAACUUUCCGCCAUACCCCACUGGCUCCAAUUUCGGACCAUAUCCUCCGGCCGGCAGUGGUCCUGCUGGUCAUGGCGCUGGUUAUCCGCCCUACAUGAAUUU (((.(((((.((((...((((((.......(((.(((......))))))........(((((((((.....)))))))))))))))..))))..))))))))......... ( -44.70) >DroPer_CAF1 3768 105 + 1 GAGCGGUGGCAACUUUCCGCCCUAUCCGACAGGUGCUAAUUUCGGACCCUAUCCACCA------GGCGGAUCGGGGGGCAAUGCCGCCGGCUAUCCGCCCUACAUGAAUUU ....(((((((..((.((.(((.(((((...((((..................)))).------..))))).))).)).)))))))))(((.....)))............ ( -37.57) >consensus UAGCGGUGGCAACUUUCCGCCAUAUCCCACUGGAGCCAAUUUCGGACCAUAUCCACCGACA___GCUGGACCGGCUGGCAAUGCAGCCGGCUAUCCGCCCUACAUGAAUUU ....(((((.......)))))...........................................(((((...((((((........))))))..)))))............ (-13.06 = -14.32 + 1.25)


| Location | 16,808,875 – 16,808,980 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.86 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16808875 105 + 23771897 AAUGUCGGACCGUACCCCCCGACACCCGCUGGUCCAGCUGGC---GGAUCCGGAUAUCCGGCCUACCCGAAUUUCAUACAACCAACCGCCGGUGGGUACCCACCAGCU ..((((((..........))))))...((((((...((((((---((..((((....)))).......(.....)..........))))))))((....)))))))). ( -40.30) >DroSec_CAF1 991 105 + 1 AACGUCGGACCGUAUCCCCCGACACCCGCUGGUCCAGCUGGU---GGAUCCGGCUAUCCGCCCUACAUGAAUUUCCCACAACCAACCGCCGCUGGGUACCCACCAGCA ...(((((..........)))))....(((((((((((.(((---((....(((.....))).....((.......)).......))))))))))......)))))). ( -36.10) >DroSim_CAF1 874 105 + 1 AACGUCGGACCGUAUCCCCCGACACCCGCUGGUCCAGCUGGU---GGAUCCGGCUAUCCGCCCUACAUGAAUUUCCCACAACCAACAGCCGGUGGGUACCCACCAGCA ...(((((..........)))))(((....)))...((((((---((....(((.....)))............(((((...(....)...)))))...)))))))). ( -35.30) >DroEre_CAF1 920 105 + 1 AGCUUCGGACCGUAUCCCCCGACACCCGCUGGUCCAGCCGGC---GGAUCCGGCUAUCAGCCAUACAUGAAUUUCCCGCAACCAACCGCUGGUGGAUACCCACCAGCA .((.((((..........))))...(((((((.....)))))---))....(((.....)))...............))........((((((((....)))))))). ( -40.20) >DroYak_CAF1 905 105 + 1 AACUUCGAACCGUAUCCCCCGACACCUGCUGGUCCAGCUGGC---GGAUCCGGCUAUCCGCCGUACAUGAAUUUCCCGCAACCUACAGCGGGUGGUUACCCACCAGCA ...........................((((((......(((---((((......))))))).......(((..(((((........)))))..)))....)))))). ( -33.40) >DroAna_CAF1 1077 105 + 1 AAUUUCGGACCAUAUCCUCCGGCCGGCAGUGGUCCUGCUGGUCAUGGCGCUGGUUAUCCGCCCUACAUGAAUUUCCCACAACCAU---CCGGUGGCUAUCCACCAGCU ......(((........)))(((((((((.....))))))))).....((((((((.(((((....(((.............)))---..))))).))...)))))). ( -33.42) >consensus AACGUCGGACCGUAUCCCCCGACACCCGCUGGUCCAGCUGGC___GGAUCCGGCUAUCCGCCCUACAUGAAUUUCCCACAACCAACCGCCGGUGGGUACCCACCAGCA ....((((((((.................))))))((((((........)))))).............)).................((.(((((....))))).)). (-23.56 = -23.86 + 0.31)

| Location | 16,808,912 – 16,809,016 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -22.42 |
| Energy contribution | -24.17 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16808912 104 - 23771897 AUUUUUGGAUAAUAAUCUAA--AACUUACAUAUCCCGCAGCUGGUGGGUACCCACCGGCGGUUGGUUGUAUGAAAUUCGGGUAGGCCGGAUAUCCGGAUCC---GCCAG ..((((((((....))))))--))....(((((((.((.((((((((....)))))))).)).))..))))).......((..((((((....)))).)).---.)).. ( -40.60) >DroSec_CAF1 1028 104 - 1 ACUUUUAGAUUAUAAUCUAC--AACUUACAUAUCCCGCUGCUGGUGGGUACCCAGCGGCGGUUGGUUGUGGGAAAUUCAUGUAGGGCGGAUAGCCGGAUCC---ACCAG ...............(((((--((((........((((((((((.(....)))))))))))..)))))))))............((.((((......))))---.)).. ( -36.40) >DroSim_CAF1 911 99 - 1 ACUUUU-----AUAAACUAC--AACUUACAUAUCCCGCUGCUGGUGGGUACCCACCGGCUGUUGGUUGUGGGAAAUUCAUGUAGGGCGGAUAGCCGGAUCC---ACCAG ......-----.....((((--(.((((((...((.((.((((((((....)))))))).)).)).)))))).......)))))((.((((......))))---.)).. ( -36.20) >DroEre_CAF1 957 103 - 1 G-UUUAGGAUUAUGAUCUAC--AACUUACAUAUCCCGCUGCUGGUGGGUAUCCACCAGCGGUUGGUUGCGGGAAAUUCAUGUAUGGCUGAUAGCCGGAUCC---GCCGG .-....(((((((((.....--..((..((...((.(((((((((((....))))))))))).)).))..))....))))...((((.....)))))))))---..... ( -38.10) >DroYak_CAF1 942 103 - 1 U-UUUAGGAUUUUGAUCAAC--AACUUACAUAUCCCGCUGCUGGUGGGUAACCACCCGCUGUAGGUUGCGGGAAAUUCAUGUACGGCGGAUAGCCGGAUCC---GCCAG .-.....(((....)))...--....(((((.((((((.((((((((((....)))))))...))).)))))).....))))).(((((((......))))---))).. ( -38.30) >DroAna_CAF1 1114 103 - 1 AUACUCCA---AUAAGAUACGAAUCAUACAUAGCCCGCAGCUGGUGGAUAGCCACCGG---AUGGUUGUGGGAAAUUCAUGUAGGGCGGAUAACCAGCGCCAUGACCAG ....(((.---....(((....)))...(((((((.....(((((((....)))))))---..))))))))))...(((((..(.((((....)).)).)))))).... ( -31.30) >consensus A_UUUUGGAUUAUAAUCUAC__AACUUACAUAUCCCGCUGCUGGUGGGUACCCACCGGCGGUUGGUUGUGGGAAAUUCAUGUAGGGCGGAUAGCCGGAUCC___ACCAG ........................((((((...((.((.((((((((....)))))))).)).)).))))))............(((.....))).............. (-22.42 = -24.17 + 1.75)


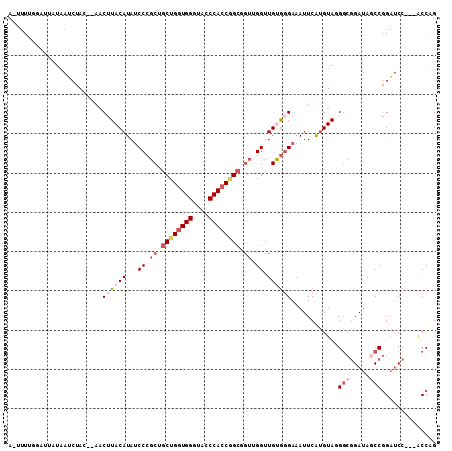
| Location | 16,808,946 – 16,809,040 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16808946 94 + 23771897 CAUACAACCAACCGCCGGUGGGUACCCACCAGCUGCGGGAUAUGUAAGUU--UUAGAUUAUUAUCCAAAAAUCGAC--UUCUU---AAUGUUUAAAGCUUA ((((...((..(.((.(((((....))))).)).)..)).))))((((((--((((((.((((.............--....)---))))))))))))))) ( -24.53) >DroSec_CAF1 1062 94 + 1 CCCACAACCAACCGCCGCUGGGUACCCACCAGCAGCGGGAUAUGUAAGUU--GUAGAUUAUAAUCUAAAAGUCGAC--UUCUU---AAAAUCUGAAGUUUA ...(((((((.((((.(((((.......))))).))))....))...)))--))((((....)))).......(((--(((..---.......)))))).. ( -25.10) >DroSim_CAF1 945 89 + 1 CCCACAACCAACAGCCGGUGGGUACCCACCAGCAGCGGGAUAUGUAAGUU--GUAGUUUAU-----AAAAGUCGAC--UUCUU---AAAAUCUGAAAUUUA .......((..(.((.(((((....))))).)).)..)).....((((((--.(((.((.(-----(((((....)--)).))---).)).))).)))))) ( -18.70) >DroEre_CAF1 991 87 + 1 CCCGCAACCAACCGCUGGUGGAUACCCACCAGCAGCGGGAUAUGUAAGUU--GUAGAUCAUAAUCCUAAA-CCUGC--UUGUU---AUAGU------UUUC (((((........((((((((....)))))))).)))))...(((((...--((((..............-.))))--...))---)))..------.... ( -25.86) >DroYak_CAF1 976 87 + 1 CCCGCAACCUACAGCGGGUGGUUACCCACCAGCAGCGGGAUAUGUAAGUU--GUUGAUCAAAAUCCUAAA-ACAGC--UUGUU---AUAGU------UUUC (((((........((.(((((....))))).)).))))).....((((((--(((..............)-)))))--)))..---.....------.... ( -24.54) >DroAna_CAF1 1151 91 + 1 CCCACAACCAU---CCGGUGGCUAUCCACCAGCUGCGGGCUAUGUAUGAUUCGUAUCUUAU---UGGAGUAUAGAAUUUUGUUCAUAUAUU-U---GUUUU ...((((...(---(((((((....)))))((.(((((((.......).)))))).))...---.)))(((((((((...)))).))))))-)---))... ( -17.30) >consensus CCCACAACCAACCGCCGGUGGGUACCCACCAGCAGCGGGAUAUGUAAGUU__GUAGAUUAUAAUCCAAAA_UCGAC__UUCUU___AAAGU_U___GUUUA ...(((.((..(.((.(((((....))))).)).)..))...)))........................................................ (-12.59 = -12.87 + 0.28)

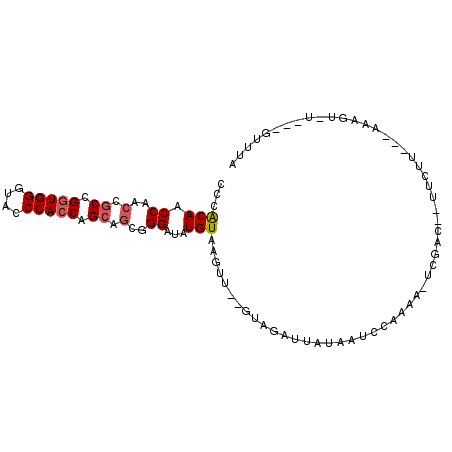
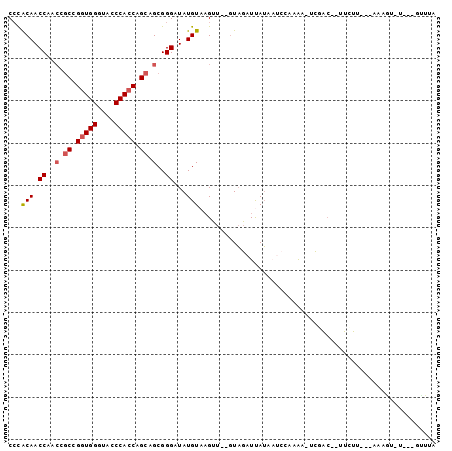
| Location | 16,808,946 – 16,809,040 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -19.26 |
| Energy contribution | -20.52 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16808946 94 - 23771897 UAAGCUUUAAACAUU---AAGAA--GUCGAUUUUUGGAUAAUAAUCUAA--AACUUACAUAUCCCGCAGCUGGUGGGUACCCACCGGCGGUUGGUUGUAUG ...(((((.......---..)))--))....((((((((....))))))--))..((((...((.((.((((((((....)))))))).)).)).)))).. ( -28.50) >DroSec_CAF1 1062 94 - 1 UAAACUUCAGAUUUU---AAGAA--GUCGACUUUUAGAUUAUAAUCUAC--AACUUACAUAUCCCGCUGCUGGUGGGUACCCAGCGGCGGUUGGUUGUGGG ...(((((.......---..)))--))..................((((--((((........((((((((((.(....)))))))))))..)))))))). ( -30.90) >DroSim_CAF1 945 89 - 1 UAAAUUUCAGAUUUU---AAGAA--GUCGACUUUU-----AUAAACUAC--AACUUACAUAUCCCGCUGCUGGUGGGUACCCACCGGCUGUUGGUUGUGGG ((((..((.(((((.---...))--)))))..)))-----)....((((--((((.(((.........((((((((....))))))))))).)))))))). ( -29.80) >DroEre_CAF1 991 87 - 1 GAAA------ACUAU---AACAA--GCAGG-UUUAGGAUUAUGAUCUAC--AACUUACAUAUCCCGCUGCUGGUGGGUAUCCACCAGCGGUUGGUUGCGGG ....------.....---.....--(((((-((..((((....))))..--)))))......((.(((((((((((....))))))))))).))..))... ( -28.80) >DroYak_CAF1 976 87 - 1 GAAA------ACUAU---AACAA--GCUGU-UUUAGGAUUUUGAUCAAC--AACUUACAUAUCCCGCUGCUGGUGGGUAACCACCCGCUGUAGGUUGCGGG ....------.(((.---(((..--...))-).)))(((....)))...--...........(((((.((((((((((....)))))))...))).))))) ( -24.40) >DroAna_CAF1 1151 91 - 1 AAAAC---A-AAUAUAUGAACAAAAUUCUAUACUCCA---AUAAGAUACGAAUCAUACAUAGCCCGCAGCUGGUGGAUAGCCACCGG---AUGGUUGUGGG .....---.-....(((((.....((..(((......---)))..)).....))))).....((((((((((((((....)))))..---..))))))))) ( -22.60) >consensus UAAAC___A_ACUAU___AACAA__GUCGA_UUUUGGAUUAUAAUCUAC__AACUUACAUAUCCCGCUGCUGGUGGGUACCCACCGGCGGUUGGUUGUGGG .....................................................((((((...((.((.((((((((....)))))))).)).)).)))))) (-19.26 = -20.52 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:14 2006