
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,798,029 – 16,798,169 |
| Length | 140 |
| Max. P | 0.998966 |

| Location | 16,798,029 – 16,798,144 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -32.12 |
| Energy contribution | -31.92 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
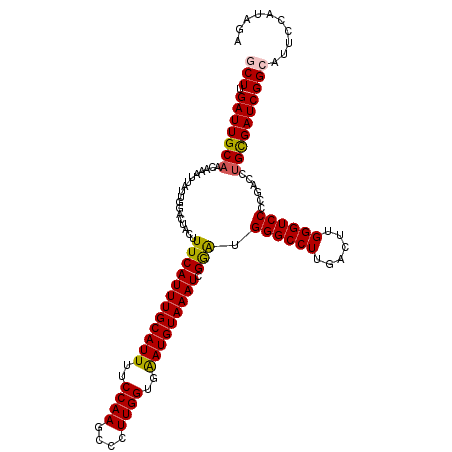
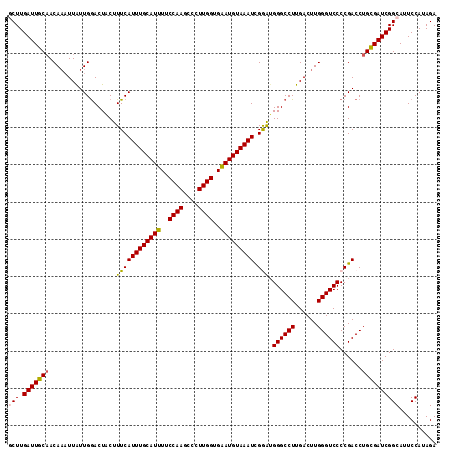
>3L_DroMel_CAF1 16798029 115 - 23771897 GCUUGAUUGCAACAAAUUAUUGGACUGCUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGACUUGGGUCCCCGACCUGCGAUCGGCAUUCCAUAGA (((.(((((((......................(((((((((.((((....)))).)))))))))((((..((((((......))))))))))..)))))))))).......... ( -37.90) >DroSec_CAF1 46366 115 - 1 GCUUGAUUGCAACAAAUUAUUGGACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGACUUGGGUCCCCGACCUGCGAUCGGCAUUCCAUAGA (((.(((((((......................(((((((((.((((....)))).)))))))))((((..((((((......))))))))))..)))))))))).......... ( -37.90) >DroSim_CAF1 51274 115 - 1 GCUUGAUUGCAACAAAUUAUUGGACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGACUUGGGUCCCCGACCUGCGAUCGGCAUUCCAUAGA (((.(((((((......................(((((((((.((((....)))).)))))))))((((..((((((......))))))))))..)))))))))).......... ( -37.90) >DroEre_CAF1 46625 115 - 1 ACUUGAUUGCAACAAAUUAUUGGACUGCUUUCAUUUGCAUUUUCCAAGCCCUUGGUGGAUGUAAAUUGAAAGGGCCUUGACUUGGGUCCCCGACCUGUGAUCGGCAUUCCACAGA ..(((((..((.(((.(((..((.((.((((((((((((((..((((....))))..)))))))).)))))))))).))).)))((((...))))))..)))))........... ( -34.10) >DroYak_CAF1 47080 115 - 1 ACUUGAUUGCUACACUUUGUUGAACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGGAUGUAAAUUGAGUGGGCCUUGACUUGGGUCCCCGACCUGCGAUCGGCAUUCCAAAGA .(((((((((........((..(.((((((..(((((((((..((((....))))..))))))))).))))))...)..))..(((((...)))))))))))((....)).))). ( -35.50) >consensus GCUUGAUUGCAACAAAUUAUUGGACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGACUUGGGUCCCCGACCUGCGAUCGGCAUUCCAUAGA (((.(((((((..................((((((((((((..((((....))))..))))))))).))).((((((......))))))......)))))))))).......... (-32.12 = -31.92 + -0.20)

| Location | 16,798,064 – 16,798,169 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 96.85 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 1.01 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
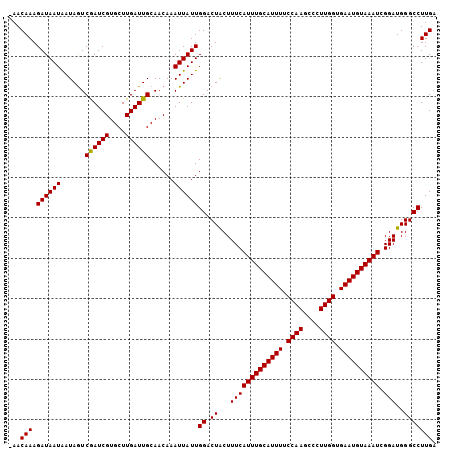
>3L_DroMel_CAF1 16798064 105 - 23771897 -AACAAAGAUAAUGAUAAUCGAUCGUGCUUGAUUGCAACAAAUUAUUGGACUGCUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGA -..(((.((((((......((((((....))))))......))))))((.((...(((((((((((((.((((....)))).)))))))))).)))..))))))). ( -24.30) >DroSec_CAF1 46401 105 - 1 -AACAAAGAUAAUAAUAGUCAAUCGUGCUUGAUUGCAACAAAUUAUUGGACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGA -..(((.((((((......((((((....))))))......))))))((.((...(((((((((((((.((((....)))).)))))))))).)))..))))))). ( -24.60) >DroSim_CAF1 51309 106 - 1 AAACAAAGAUAAUAAUAGUCGAUCGUGCUUGAUUGCAACAAAUUAUUGGACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGA ...(((.((((((......((((((....))))))......))))))((.((...(((((((((((((.((((....)))).)))))))))).)))..))))))). ( -24.30) >consensus _AACAAAGAUAAUAAUAGUCGAUCGUGCUUGAUUGCAACAAAUUAUUGGACUACUUUCAUUUGCAUUUUCCAAGCCCUUGGUGAAUGUAAAUCGGAUGGGCCUUGA ...(((.((((((......((((((....))))))......))))))((.((...(((((((((((((.((((....)))).)))))))))).)))..))))))). (-24.62 = -24.40 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:09 2006