
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,791,588 – 16,791,722 |
| Length | 134 |
| Max. P | 0.980237 |

| Location | 16,791,588 – 16,791,685 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
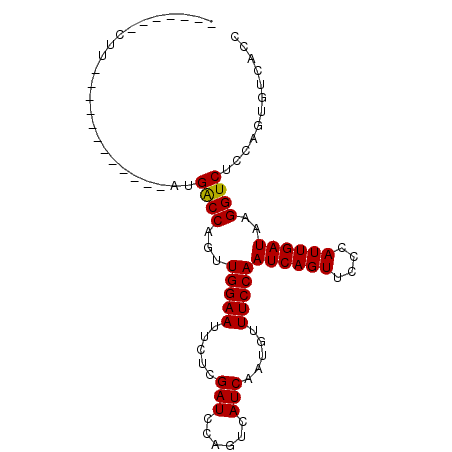
>3L_DroMel_CAF1 16791588 97 - 23771897 GCAUGCACUUCACUGCAAAUAAAUGGCCAAUUGGAAUUCUCGAUCCAGUCAUCAAUAUUUCCAAUCAGUUUCCAUUGAUAAGGUCUCCAGUGUCACC (((((((......)))........((((...(((((.....(((......))).....)))))((((((....))))))..))))....)))).... ( -16.80) >DroSec_CAF1 40037 78 - 1 -------CUU------------AUGACCAGUUGGAAUUCUCGAUCCAGUCAUCGAUGUUUCCAAUCAGUUCCCAUUGAUAAGGUCUCCAGUGUCACC -------...------------..((((...(((((...(((((......)))))...)))))((((((....))))))..))))............ ( -18.50) >DroSim_CAF1 44393 78 - 1 -------CUU------------AUGACCAGUUGGAAUUCUCGAUCCAGUCAUCUAUGUUUCCAAUCAGUUCCCAUUGAUAAGGUCUCCAGUGUCACC -------...------------..((((...(((((.....(((......))).....)))))((((((....))))))..))))............ ( -13.90) >consensus _______CUU____________AUGACCAGUUGGAAUUCUCGAUCCAGUCAUCAAUGUUUCCAAUCAGUUCCCAUUGAUAAGGUCUCCAGUGUCACC ........................((((...(((((.....(((......))).....)))))((((((....))))))..))))............ (-13.92 = -13.70 + -0.22)


| Location | 16,791,619 – 16,791,722 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.41 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
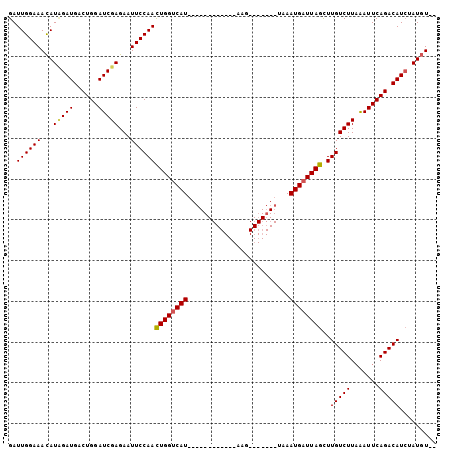
>3L_DroMel_CAF1 16791619 103 + 23771897 GAUUGGAAAUAUUGAUGACUGGAUCGAGAAUUCCAAUUGGCCAUUUAUUUGCAGUGAAGUGCAUGCUAAAUGAUUAGUUUGUCUUUGAUUCAGACAUCUAUUU-- .............((((.(((((((((..((...(((((..((((((..((((......))))...))))))..))))).))..))))))))).)))).....-- ( -25.50) >DroSec_CAF1 40068 86 + 1 GAUUGGAAACAUCGAUGACUGGAUCGAGAAUUCCAACUGGUCAU------------AAG-------UAAAUGAUUAGCUUGUCUUAAAUUCAGACAUAUAUGUGC ..((((((...(((((......)))))...))))))((((((((------------...-------...))))))))..(((((.......)))))......... ( -21.10) >DroSim_CAF1 44424 84 + 1 GAUUGGAAACAUAGAUGACUGGAUCGAGAAUUCCAACUGGUCAU------------AAG-------UAAAUGAUUAGCUUGUCUUAAAUUCAGACAUCUAUGU-- ........(((((((((.((((((.((((....(((((((((((------------...-------...)))))))).)))))))..)))))).)))))))))-- ( -26.50) >consensus GAUUGGAAACAUAGAUGACUGGAUCGAGAAUUCCAACUGGUCAU____________AAG_______UAAAUGAUUAGCUUGUCUUAAAUUCAGACAUCUAUGU__ ..((((((...(((((......)))))...))))))((((((((.........................))))))))..(((((.......)))))......... (-14.07 = -14.41 + 0.34)


| Location | 16,791,619 – 16,791,722 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -11.62 |
| Energy contribution | -11.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
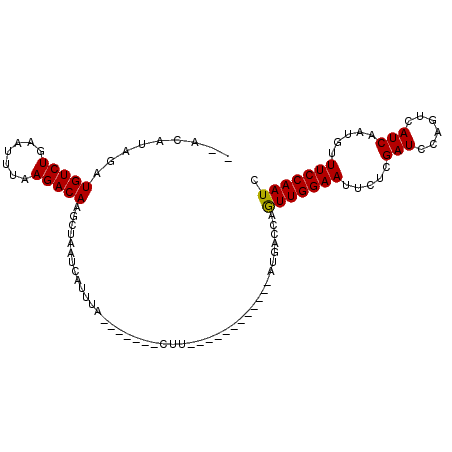
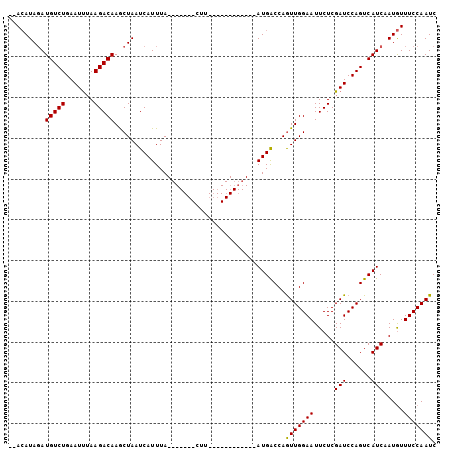
>3L_DroMel_CAF1 16791619 103 - 23771897 --AAAUAGAUGUCUGAAUCAAAGACAAACUAAUCAUUUAGCAUGCACUUCACUGCAAAUAAAUGGCCAAUUGGAAUUCUCGAUCCAGUCAUCAAUAUUUCCAAUC --.....((((.(((.(((.............(((((((...((((......))))..)))))))((....)).......))).))).))))............. ( -20.90) >DroSec_CAF1 40068 86 - 1 GCACAUAUAUGUCUGAAUUUAAGACAAGCUAAUCAUUUA-------CUU------------AUGACCAGUUGGAAUUCUCGAUCCAGUCAUCGAUGUUUCCAAUC .........(((((.......)))))((((..((((...-------...------------))))..))))((((...(((((......)))))...)))).... ( -17.70) >DroSim_CAF1 44424 84 - 1 --ACAUAGAUGUCUGAAUUUAAGACAAGCUAAUCAUUUA-------CUU------------AUGACCAGUUGGAAUUCUCGAUCCAGUCAUCUAUGUUUCCAAUC --(((((((((.(((...(..(((..((((..((((...-------...------------))))..)))).....)))..)..))).)))))))))........ ( -21.30) >consensus __ACAUAGAUGUCUGAAUUUAAGACAAGCUAAUCAUUUA_______CUU____________AUGACCAGUUGGAAUUCUCGAUCCAGUCAUCAAUGUUUCCAAUC .........(((((.......)))))..........................................(((((((.....(((......))).....))))))). (-11.62 = -11.40 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:06 2006