
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,788,280 – 16,788,382 |
| Length | 102 |
| Max. P | 0.935339 |

| Location | 16,788,280 – 16,788,382 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16788280 102 - 23771897 AGAUCGAGAAGGAAAAAA---ACGAAAAAUAAUUCGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAAAUCGGUGAACGGUGGUA-CAGCUGCUGUAUACCGGUGG .((((.....(.......---.)......(((((.((((....)))).)))))...))))........(((((((.((((..(..-...)..)))).))))))).. ( -28.30) >DroSec_CAF1 36808 103 - 1 AGAUCGAGAAGGAAAAAAA--AUGAAAAAUAAUUCGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAAAUCGGUGAACGGUGGAA-CAGCUGCUGUAUACCGGUGG .((((..............--.(....).(((((.((((....)))).)))))...))))........(((((((.((((..(..-...)..)))).))))))).. ( -26.60) >DroSim_CAF1 41127 105 - 1 AGAUCGAGAAGGAAAAAAAAGACGAAAAAUAAUUCGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAAAUCGGUGAACGGUGGAA-CAGCUGCUGUAUACCGGUGG .((((.....(...........)......(((((.((((....)))).)))))...))))........(((((((.((((..(..-...)..)))).))))))).. ( -26.40) >DroEre_CAF1 37247 101 - 1 AGAUCGAGAAG-AAAAAA---ACGAAAAAUAAUUUGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAAAGCGGUGAAUGGUGGAA-CAGCUGCUGUAUACCGGUGG .((((.....(-......---.)......(((((.((((....)))).)))))...))))(((.....)))((((.((((..(..-...)..)))).))))..... ( -21.90) >DroYak_CAF1 37432 102 - 1 AGAUCGAGAAGGAAAAAA---ACGAAAAAUAAUUCGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAAAGCGGUGAACGGUGGAA-CAGCUGCUGUAUACCGGUGG .((((.....(.......---.)......(((((.((((....)))).)))))...))))(((.....)))((((.((((..(..-...)..)))).))))..... ( -26.00) >DroAna_CAF1 36222 85 - 1 ----------GGA-----------AAUAAUAAUUUGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAGAAAGGUGAACGGCGGAGCCAGCUGCUGUAUGCCGCUUG ----------.((-----------..((.(((((.((((....)))).))))).))..))...........((((.(((((((......))))))).))))..... ( -23.00) >consensus AGAUCGAGAAGGAAAAAA___ACGAAAAAUAAUUCGCCAAAAAUGGCCAAUUAAUAGAUCGCUAUCAAAUCGGUGAACGGUGGAA_CAGCUGCUGUAUACCGGUGG .............................(((((.((((....)))).)))))((((....)))).....(((((.(((((((......))))))).))))).... (-21.36 = -21.12 + -0.25)

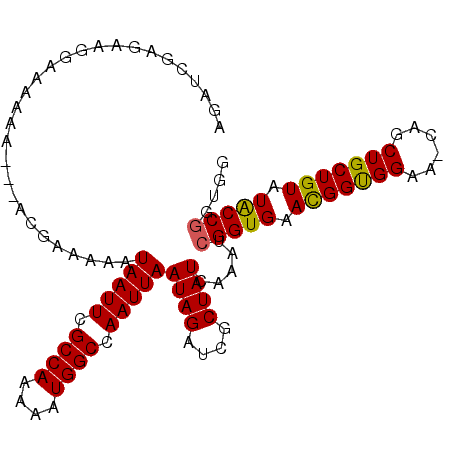
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:04 2006