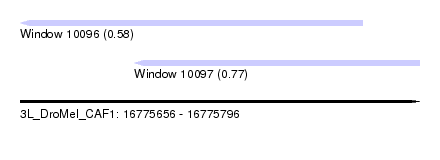
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,775,656 – 16,775,796 |
| Length | 140 |
| Max. P | 0.771829 |

| Location | 16,775,656 – 16,775,776 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.03 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
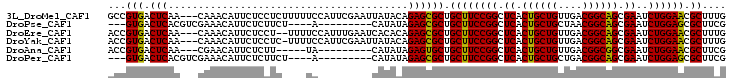
>3L_DroMel_CAF1 16775656 120 - 23771897 CUCCUCUUUUUCCAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCACAAGAAGCAGGAGCGACACAACAAGAUGCU (((((((((((..............(((((((....((((((.((.((((((....)))))).))..))))))))))))).((....))..)))))).)))))................. ( -37.80) >DroVir_CAF1 23839 109 - 1 CUCU-----------CUCUGCUGUAUAGAGCGCCGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAGCGGUUCGCGGAGAUACACAAGAAACAGGAGCGUCAGCACAAGAUGCU ...(-----------((((((.((.....))((((((((.(..((.((((((....)))))).))..).))))))))..)))))))...............((((((.......)))))) ( -42.40) >DroPse_CAF1 24812 107 - 1 CUCUUCU----A---------CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUAACGGCAGCGAAUCUGGAGCGCUUCGCGGAGAUCCACAAGAAGCAGGAGCGGCACAACAAGAUGUU ((((...----.---------.....)))).((((((((..((((..((((((.....))))))......))))(((((..((....))....))))).))))))))............. ( -38.00) >DroGri_CAF1 24567 109 - 1 UUCU-----------UGGACAAUUACAGAGCGCCGCUUCUGGUUCACUGCUGCUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCACAAGAAACAGGAGCGUCAGCACAAAAUGAU ((((-----------((........(((((((...((...(((((.((((((....)))))).))))).))..))))))).((....)).))))))......((....)).......... ( -34.50) >DroAna_CAF1 24496 106 - 1 CUCUU-----UA---------CAUAUAGAGUGCUGCUUCCGGCUCACUGCUGUUGACGGCGGCGAAUCUGGAACGCUUCGCGGAGAUCCACAAGAAGCAGGAGCGCCGCAACAAGAUGCU .....-----..---------......(.(((((.(((((((.((.((((((....)))))).))..)))))..(((((..((....))....))))))).))))))(((......))). ( -36.00) >DroPer_CAF1 25118 107 - 1 CUCUUCU----A---------CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAGCGCUUCGCGGAGAUCCACAAGAAGCAGGAGCGGCACAACAAGAUGUU ((((...----.---------.....)))).((((((((..((((..((((((.....))))))......))))(((((..((....))....))))).))))))))............. ( -38.00) >consensus CUCUU______A_________CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAACGCUUCGCGGAGAUCCACAAGAAGCAGGAGCGGCACAACAAGAUGCU ((((.........................(((..((((((((.((.((((((....)))))).))..)))))).))..)))((....))..........))))................. (-26.86 = -26.03 + -0.83)


| Location | 16,775,696 – 16,775,796 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16775696 100 - 23771897 GCCGUGACUCAA---CAAACAUUCUCCUCUUUUUCCAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUG ...((......)---)................................(((((((....((((((.((.((((((....)))))).))..))))))))))))) ( -25.80) >DroPse_CAF1 24852 87 - 1 ---GUGACUCACGUCGAAACAUUCUCUUCU----A---------CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUAACGGCAGCGAAUCUGGAGCGCUUCG ---.((((....))))..............----.---------......((((((....(((((.((.((((((....)))))).))..))))))))))).. ( -26.30) >DroEre_CAF1 24937 98 - 1 ACCGUGACUCAA---CAAACAUUCUCCU--UUUUCCAUUUGAAUCACACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUG ...((((.((((---.............--........)))).)))).(((((((....((((((.((.((((((....)))))).))..))))))))))))) ( -30.90) >DroYak_CAF1 24701 99 - 1 ACCGUGACUCAA---CAAACAUUCUCCUC-UUUUCCAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUG ...((......)---).............-..................(((((((....((((((.((.((((((....)))))).))..))))))))))))) ( -25.80) >DroAna_CAF1 24536 86 - 1 ACCGUGACUCAA---CGAACAUUCUCUU-----UA---------CAUAUAGAGUGCUGCUUCCGGCUCACUGCUGUUGACGGCGGCGAAUCUGGAACGCUUCG ..(((......)---))..((((((.(.-----..---------...).))))))..((((((((.((.((((((....)))))).))..)))))).)).... ( -24.70) >DroPer_CAF1 25158 87 - 1 ---GUGACUCACGUCGAAACAUUCUCUUCU----A---------CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAGCGCUUCG ---(((.(((..((.(((........))).----)---------)...((((.(((((((..((((........))))..)))))))..)))))))))).... ( -28.20) >consensus ACCGUGACUCAA___CAAACAUUCUCCUC____UA_________CAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUCG ...(((.(((........................................)))))).((((((((.((.((((((....)))))).))..)))))).)).... (-22.68 = -22.18 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:56 2006