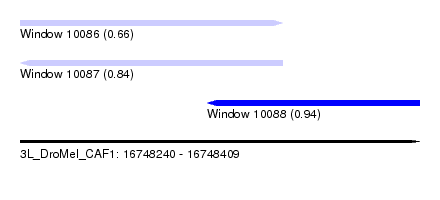
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,748,240 – 16,748,409 |
| Length | 169 |
| Max. P | 0.944609 |

| Location | 16,748,240 – 16,748,351 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -24.02 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16748240 111 + 23771897 CGGUUUAUAGCAACUGCAUGCGUAGUGCGACAGAGAGUUGCUAUAAAAAUAUAUUGGAACGUCAACAAACAUAUGCCACGAAAGAAAAUAUUCCGAUUCAACGGAAAAUGA .((((((((((((((...((((.....)).))...))))))))))).......(((.........)))......))).(....)......(((((......)))))..... ( -25.40) >DroSim_CAF1 1085 111 + 1 CGGUUUAUAGCAACUGCAUGCGUAGUGCGACAGAGAGUUGCUAUAAAAGUAUAUUGGAACGUCAACAAACAUAUGCCACGAAAGAAAAUAUUCCGAUUCGACGGAAUAUGA .((((((((((((((...((((.....)).))...)))))))))))..((.(.((((....)))).).))....))).(....)...((((((((......)))))))).. ( -29.10) >DroYak_CAF1 1100 111 + 1 CGCUUUAUAGCAGCUGCAUGCGUAGUGCGACACCGAGUUGCUAUAAAAAUAUAUUGGAACGUCAACAAACAUAUGCCACGAAAGAAAAUAUUCCGAUUCGACGGAACAUGA ...((((((((((((((((.....))))(....).))))))))))))........................((((...(....)......(((((......))))))))). ( -23.70) >consensus CGGUUUAUAGCAACUGCAUGCGUAGUGCGACAGAGAGUUGCUAUAAAAAUAUAUUGGAACGUCAACAAACAUAUGCCACGAAAGAAAAUAUUCCGAUUCGACGGAAAAUGA ...((((((((((((...((((.....)).))...))))))))))))......................(((......(....)......(((((......))))).))). (-24.02 = -23.80 + -0.22)

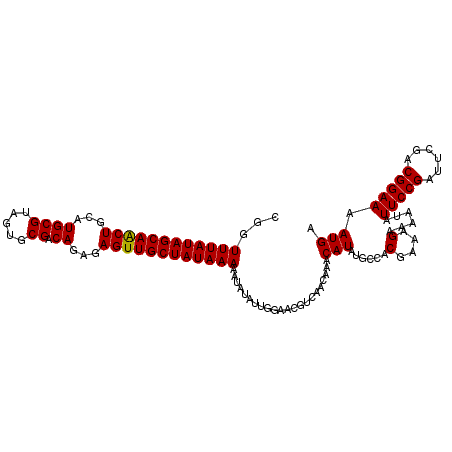
| Location | 16,748,240 – 16,748,351 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16748240 111 - 23771897 UCAUUUUCCGUUGAAUCGGAAUAUUUUCUUUCGUGGCAUAUGUUUGUUGACGUUCCAAUAUAUUUUUAUAGCAACUCUCUGUCGCACUACGCAUGCAGUUGCUAUAAACCG .....(((((......)))))............(((...(((((....))))).))).......((((((((((((.(.((.((.....)))).).))))))))))))... ( -24.80) >DroSim_CAF1 1085 111 - 1 UCAUAUUCCGUCGAAUCGGAAUAUUUUCUUUCGUGGCAUAUGUUUGUUGACGUUCCAAUAUACUUUUAUAGCAACUCUCUGUCGCACUACGCAUGCAGUUGCUAUAAACCG ..((((((((......)))))))).........(((...(((((....))))).))).......((((((((((((.(.((.((.....)))).).))))))))))))... ( -27.90) >DroYak_CAF1 1100 111 - 1 UCAUGUUCCGUCGAAUCGGAAUAUUUUCUUUCGUGGCAUAUGUUUGUUGACGUUCCAAUAUAUUUUUAUAGCAACUCGGUGUCGCACUACGCAUGCAGCUGCUAUAAAGCG ..((((((((......)))))))).........(((...(((((....))))).)))......((((((((((.((((((((......)))).)).)).)))))))))).. ( -27.10) >consensus UCAUAUUCCGUCGAAUCGGAAUAUUUUCUUUCGUGGCAUAUGUUUGUUGACGUUCCAAUAUAUUUUUAUAGCAACUCUCUGUCGCACUACGCAUGCAGUUGCUAUAAACCG ..((((((((......)))))))).........(((...(((((....))))).))).......((((((((((((...((.((.....))))...))))))))))))... (-25.61 = -26.17 + 0.56)



| Location | 16,748,319 – 16,748,409 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.99 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16748319 90 - 23771897 GGUUAAAUGUACGAUCGAAUGGCUUUCUGAAUAUCUGCCAGUUCGGUCUUUAAUUAAUUCAUUUUCCGUUGAAUCGGAAUAUUUUCUUUC ....((((((..(((((((((((.............)))).)))))))................((((......))))))))))...... ( -19.52) >DroSim_CAF1 1164 90 - 1 AGUUAAAUGUACGAUCGAAUGGCUUUCUGGAUGUUUGCCAGUUCGGUCUUUAAUUAAUUCAUAUUCCGUCGAAUCGGAAUAUUUUCUUUC (((((((.....(((((((((((...(.....)...)))).)))))))))))))).....((((((((......))))))))........ ( -23.10) >DroYak_CAF1 1179 90 - 1 GGUUAAAUGGCUGAUCGAAUGGCUGUCUGAAUCUCUGCCAGUCCGAUCUGUAAUCAAUUCAUGUUCCGUCGAAUCGGAAUAUUUUCUUUC (((((.......(((((..((((.............))))...)))))..))))).....((((((((......))))))))........ ( -21.92) >consensus GGUUAAAUGUACGAUCGAAUGGCUUUCUGAAUAUCUGCCAGUUCGGUCUUUAAUUAAUUCAUAUUCCGUCGAAUCGGAAUAUUUUCUUUC (((((((.....(((((((((((.............)))).)))))))))))))).....((((((((......))))))))........ (-20.76 = -20.99 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:47 2006