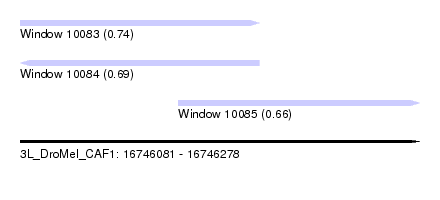
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,746,081 – 16,746,278 |
| Length | 197 |
| Max. P | 0.739744 |

| Location | 16,746,081 – 16,746,199 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -21.63 |
| Energy contribution | -22.31 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16746081 118 + 23771897 CCAGCAAAACCACCCACCCACUACUGGCCCCACACACACUUGACCCCAAGGGGAACUUGGCCCCUAUGCAGAGCCUUUAUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACA ...................(((..((((.((............(((....)))...((((((((.(((.((...)).)))))))))))...)).))))..)))............... ( -30.40) >DroSec_CAF1 152 114 + 1 -CUGCCCUG---CCCACCCACUACUUGUCCCACACACACUUGACCCCAAGGGGAACUUGGCCCCUAUGCAGAGCCUUUAUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACA -..((((((---....(((......(((.....)))..((((....)))))))...((((((((.(((.((...)).))))))))))).)))).)).((((..........))))... ( -28.80) >DroSim_CAF1 158 115 + 1 CCAGCAAAA---CCCACCCACUACUGGCCCCACACACACUUGACCCCAAGGGGAACUUGGCCCCUAUGCAGAGCCUUAAUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACA .........---.......(((..((((.((............(((....)))...((((((((................))))))))...)).))))..)))............... ( -30.29) >DroEre_CAF1 376 113 + 1 CCACCAAAACCACCCACCC---ACUGGCCUCA--AACACUUGACCCCAAAGGGAACUUGGCCCGUAUGUAGAGCCUUUACGGAGCCAAACGGGCGCCGAAAGUUCAUAUUAUUUCGCA ............(((.(((---..(((..(((--(....))))..)))..)))...((((((((((...........))))).)))))..))).((.((((..........)))))). ( -31.40) >DroYak_CAF1 173 113 + 1 CGACCCAGUAAUUCCACCC---ACUGGCCCCA--CACACUUGACCCCAAAGGGAACUUGGCCCCUAUGCAGAGCCUUUACGGGGCCAAAUGAGCGCGGAAAGUUCAUAUUAUUUCGCA (((...((((((.......---..(((((((.--.........(((....))).....(((..((....)).))).....))))))).((((((.......))))))))))))))).. ( -30.10) >consensus CCAGCAAAA___CCCACCCACUACUGGCCCCACACACACUUGACCCCAAGGGGAACUUGGCCCCUAUGCAGAGCCUUUAUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACA .........................(((.((............(((....)))...((((((((................))))))))...)).)))((((..........))))... (-21.63 = -22.31 + 0.68)


| Location | 16,746,081 – 16,746,199 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -33.50 |
| Energy contribution | -34.70 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16746081 118 - 23771897 UGUGAAAUAAUGUGAACUUUCGGCGCCUAUUUGGCCCCAUAAAGGCUCUGCAUAGGGGCCAAGUUCCCCUUGGGGUCAAGUGUGUGUGGGGCCAGUAGUGGGUGGGUGGUUUUGCUGG .(..((((...(....)......((((((((((((((((((..((((((.....)))))).....(((....))).........))))))))))..))))))))....))))..)... ( -46.50) >DroSec_CAF1 152 114 - 1 UGUGAAAUAAUGUGAACUUUCGGCGCCUAUUUGGCCCCAUAAAGGCUCUGCAUAGGGGCCAAGUUCCCCUUGGGGUCAAGUGUGUGUGGGACAAGUAGUGGGUGGG---CAGGGCAG- ............((..((.((.((.((((((((..((((((..((((((.....)))))).....(((....))).........)))))).))))))).).)).))---.))..)).- ( -40.00) >DroSim_CAF1 158 115 - 1 UGUGAAAUAAUGUGAACUUUCGGCGCCUAUUUGGCCCCAUUAAGGCUCUGCAUAGGGGCCAAGUUCCCCUUGGGGUCAAGUGUGUGUGGGGCCAGUAGUGGGUGGG---UUUUGCUGG .(..((((...(....)......(((((((((((((((((...((((((.....)))))).....(((....)))..........)))))))))..)))))))).)---)))..)... ( -44.80) >DroEre_CAF1 376 113 - 1 UGCGAAAUAAUAUGAACUUUCGGCGCCCGUUUGGCUCCGUAAAGGCUCUACAUACGGGCCAAGUUCCCUUUGGGGUCAAGUGUU--UGAGGCCAGU---GGGUGGGUGGUUUUGGUGG ..((((((........(((((((.(((.....))).))).))))((.((((.(((.((((.....(((....)))((((....)--))))))).))---).)))))).)))))).... ( -35.80) >DroYak_CAF1 173 113 - 1 UGCGAAAUAAUAUGAACUUUCCGCGCUCAUUUGGCCCCGUAAAGGCUCUGCAUAGGGGCCAAGUUCCCUUUGGGGUCAAGUGUG--UGGGGCCAGU---GGGUGGAAUUACUGGGUCG ..(((...............((.(((.((((((((((((....((((((.....))))))(((....))))))))))))))).)--)).))(((((---((......))))))).))) ( -44.70) >consensus UGUGAAAUAAUGUGAACUUUCGGCGCCUAUUUGGCCCCAUAAAGGCUCUGCAUAGGGGCCAAGUUCCCCUUGGGGUCAAGUGUGUGUGGGGCCAGUAGUGGGUGGG___UUUUGCUGG .......................(((((((((((((((((...((((((.....)))))).....(((....)))..........))))))))))..))))))).............. (-33.50 = -34.70 + 1.20)



| Location | 16,746,159 – 16,746,278 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16746159 119 + 23771897 AUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACAAGGUUCGCUCUUUUGUAUGAAAACCAAAUACAACCUUUUGAAAUAGUAAAAGAAUAGCAUGGAAGCUUGUAAUUUCUAA ..((.(((.....))).))((((((((..(((((((((.(((((.........(((((.........)))))))))).)))))))))....))))(((......))).....))))... ( -27.30) >DroSec_CAF1 226 119 + 1 AUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACAAGGUUCGCUCUUUUGUACAAAAACCAAAUACAACCUUUUGAAAUAGUAAAAGAAAAGCGUGGGGCCAUGUAAUUUCUAA .(((.(((.....))).))).........(((((((((.((((((......((((.........))))...)))))).)))))))))...(((((.(((((....)))))..))))).. ( -30.00) >DroSim_CAF1 233 119 + 1 AUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACAAGGUUCGCUCUUUUGUACAAAAACCAAAUACCACCUUUUGAAAUAGAACAAGAAUAGCGUGGGGCCAUGUAAUUUCUAA .(((.(((.....))).)))...((((....(((((((.(((((.......((((.........))))....))))).))))))))))).((((..(((((....)))))...)))).. ( -29.70) >DroEre_CAF1 449 118 + 1 ACGGAGCCAAACGGGCGCCGAAAGUUCAUAUUAUUUCGCAAGGUUUGCUCUUUUGUAUACAAACCAAAUACAACCUUUUGAAAUAGUAAUCGAAU-GUUUAGGCUCUUGUGAAUUAUAA (((((((((((((..((.(....)....((((((((((.((((((......((((....))))........)))))).))))))))))..))..)-)))).))))).)))......... ( -30.84) >DroYak_CAF1 246 118 + 1 ACGGGGCCAAAUGAGCGCGGAAAGUUCAUAUUAUUUCGCAAGGUUCGCUCUUUUGUAUGAAAACCAAAUACAACCUUUUGAAAUAGUAAACGAAU-GUUUAGGGGCUUCUGAAUUCUAA ..((((((...(((((.((((....)).((((((((((.(((((.........(((((.........)))))))))).))))))))))..))...-)))))..)))))).......... ( -28.30) >consensus AUGGGGCCAAAUAGGCGCCGAAAGUUCACAUUAUUUCACAAGGUUCGCUCUUUUGUAUAAAAACCAAAUACAACCUUUUGAAAUAGUAAAAGAAUAGCGUGGGGCCUUGUAAUUUCUAA .(((.(((.....))).)))...((((..(((((((((.((((((......((((.........))))...)))))).)))))))))....))))........................ (-19.66 = -20.18 + 0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:45 2006