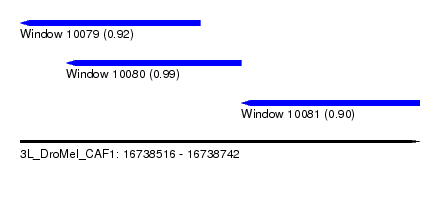
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,738,516 – 16,738,742 |
| Length | 226 |
| Max. P | 0.994413 |

| Location | 16,738,516 – 16,738,618 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.57 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16738516 102 - 23771897 UAGCUUUGAA--UCCUACCUGGAAAAUGUUGUCCCAUUCAGGUAGGCCGCGGAAAAUUCGCUCGUAUUUCCCUC--AACAUAUCCU----AGCAAUAUGGCACU------C----UUUUC ..((.((((.--.((((((((((.............))))))))))..(((((...)))))...........))--))(((((...----....)))))))...------.----..... ( -22.12) >DroGri_CAF1 42841 101 - 1 UUGCCUUGCAUUGUCUACCUGGAAAAG--CGU----CUCAGGUAAG-CGUGAAA-AUUCGCCUGUAUUAUCGUUUUG-------GC----UGCCGCAUGGCAUUUUUCUUUUUCCUUUUC .((((.(((...(..((((((((....--..)----).))))))..-)......-....(((..............)-------))----....))).)))).................. ( -24.94) >DroSim_CAF1 50150 102 - 1 UAGCUUUGAA--UCCUACCUGGAAAAUGUUGUCCCACUCAGGUAGUCCGCGGAAAAUUCGCUCGUAUUUCCCUC--AACAUAUCCC----GGCAACAUGGCACU------C----UUUUC ..(((.....--(((.....)))..((((((((.......((....))(((((...))))).............--..........----)))))))))))...------.----..... ( -19.70) >DroAna_CAF1 40095 99 - 1 U----GGAAA--UCCUACCUGGAAAAG--CGACCCAUUCAGGUAGGACGCGGAA-AUUUGCUCGAAUUUCCAUCCUGGCAUAUACCAUCCUGCCGUAUGGAG--------U----UUUUC .----((...--.))......((((((--(...((((...(((((((...((((-((((....))))))))....(((......))))))))))..)))).)--------)----))))) ( -32.90) >consensus UAGCUUUGAA__UCCUACCUGGAAAAG__CGUCCCACUCAGGUAGGCCGCGGAA_AUUCGCUCGUAUUUCCCUC__AACAUAUCCC____UGCAACAUGGCACU______C____UUUUC ..(((.((.....((((((((((.............))))))))))..(((((...)))))..................................)).)))................... (-13.82 = -14.57 + 0.75)


| Location | 16,738,542 – 16,738,641 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -16.47 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16738542 99 - 23771897 --UCCCUUUCUCUACCUGAAUCCUCUAGCUUUGAAUCCUACCUGGAAAAUGUUGUCCCAUUCAGGUAGGCCGCGGAAAAUUCGCUCGUAUUUCCCUC--AACA --........(((((((((((....((((.((...(((.....))).)).))))....)))))))))))..(((((...))))).............--.... ( -22.30) >DroSim_CAF1 50176 101 - 1 UCUCCCUUUCUCUACCUGAAUCCUCUAGCUUUGAAUCCUACCUGGAAAAUGUUGUCCCACUCAGGUAGUCCGCGGAAAAUUCGCUCGUAUUUCCCUC--AACA ...........((((((((......((((.((...(((.....))).)).))))......))))))))...(((((...))))).............--.... ( -19.00) >DroAna_CAF1 40123 92 - 1 UCUUUCUUUUUCUACCUGG----UCU----GGAAAUCCUACCUGGAAAAG--CGACCCAUUCAGGUAGGACGCGGAA-AUUUGCUCGAAUUUCCAUCCUGGCA ..................(----((.----(((..(((((((((((....--.......)))))))))))...((((-((((....)))))))).))).))). ( -31.00) >consensus UCUCCCUUUCUCUACCUGAAUCCUCUAGCUUUGAAUCCUACCUGGAAAAUGUUGUCCCAUUCAGGUAGGCCGCGGAAAAUUCGCUCGUAUUUCCCUC__AACA ...........((((((((......((((......(((.....)))....))))......))))))))...(((((...)))))................... (-16.47 = -16.53 + 0.06)


| Location | 16,738,641 – 16,738,742 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16738641 101 - 23771897 CAGAAAUUCCUAACCUAAAGGACAAGCAGCUGAUCCUGGCAAAUGGCAACGCCGGCUGCUCGCUCUCCUUUUGUCCUUGCACAAUUGCUUGAGGGUUUCCC------------- ..(((((.(((......(((((((((.((..((..(.((((..((((...))))..)))).).))..)))))))))))(((....)))...))))))))..------------- ( -28.50) >DroSim_CAF1 51037 113 - 1 CAGAAAUUCCUAACCCAAAGGACAAGCAGCUGAUCCUGGCAAAUGGCAACGCCGGCUGCUCGCUCUCCU-UUGUCCUUGCACAAUUGCUUGAGGGUUUCCCUCGCACUCUCCCU .(((...........((((((((.((((((((....((.(....).))....)))))))).)...))))-)))............(((..(((((...)))))))).))).... ( -30.30) >DroAna_CAF1 40218 100 - 1 -----CUUUCGGACC-GAAGGAUCUGCAGCUGAUCCUGGCAAAUGUCAAAGCCGGCUGCUCUC-CUCCA-UUGUCCUUGCACAAUCACUUAAGGGUUUUCCUUUCUCU------ -----.....((((.-(.((((...(((((((....((((....))))....)))))))..))-)).).-..))))..............(((((....)))))....------ ( -27.50) >consensus CAGAAAUUCCUAACC_AAAGGACAAGCAGCUGAUCCUGGCAAAUGGCAACGCCGGCUGCUCGCUCUCCU_UUGUCCUUGCACAAUUGCUUGAGGGUUUCCCU__C_CU______ ...........(((((.(((((((((((((((....((.(....).))....))))))))...........)))))))((......))....)))))................. (-21.75 = -22.53 + 0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:41 2006