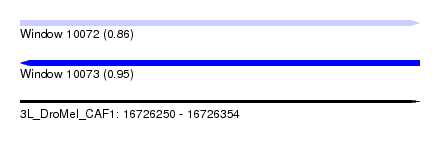
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,726,250 – 16,726,354 |
| Length | 104 |
| Max. P | 0.946004 |

| Location | 16,726,250 – 16,726,354 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
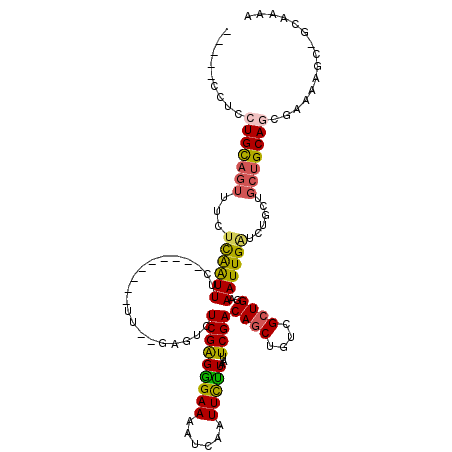
| Reading direction | forward |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -15.60 |
| Energy contribution | -17.10 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16726250 104 + 23771897 UUUUGUAGCUUUUCGCUGCAGCAGCAGAUCAAUUUCCAGCGACAGCUGUCGAAUCAAGAAUUGAUUUUCCCUCGAGACUC--AA---------GAAACUGAGAAACUGCAGGAAA----- ...((((((.....))))))((((.((((((((((....((((....))))......))))))))))..........(((--(.---------.....))))...))))......----- ( -28.20) >DroSec_CAF1 33584 90 + 1 UU--------------UGCAGCAGCAGAUCAAUUUCCAGCGACAGCUGUCGAAUCAAGAAUCGAUUUUCCCUCGAUACCC--AA---------GAAAUUGAGAAACUGCAGGAAA----- ..--------------....((((....((((((((((((....))))...........(((((.......)))))....--..---------))))))))....))))......----- ( -23.00) >DroSim_CAF1 38128 90 + 1 UU--------------UGCAGCAGCAGACCAAUUUCCAGCGACAGCUGUCGAAUCAAGAAUCGAUUUUCCCUCGAGACCC--AA---------GAAAUUGAGAAACUGCAAGAAA----- ((--------------(((((...(....(((((((((((....))))((((.....(((......)))..)))).....--..---------))))))).)...)))))))...----- ( -19.40) >DroEre_CAF1 34129 109 + 1 UUUUGCUGCUUUUCGCUGCAGCGGCAGAUCAAUUUCCAGCGACAGCUGUCGAAUUAGGAAUUGAUUUUCCCCCGAGACUA--AA---------GGAAUCAAGAAACUGCAGGAGGCGAAA .((((((.....((.((((((.((.(((((((((((...((((....)))).....))))))))))).)).((.......--..---------))..........)))))))))))))). ( -32.60) >DroYak_CAF1 37030 108 + 1 -UUUGCCGCUUUUCGCUGCAGCGGCAGAUCAAUUUUCAGCGACAGCUGUCGAAUUAAGAAUUGAUUUUCCCUCGAGACUA--AA---------GGAAUCGAGAAACUGCAGGAGGCGAAA -((((((.....((.((((((.((.(((((((((((...((((....)))).....))))))))))).))(((((..(..--..---------.)..)))))...)))))))))))))). ( -38.00) >DroAna_CAF1 29130 120 + 1 UUUUGCAUCUUUUCACUGCGCCAGCAGAUCAAUUUCCAGCGACAGUUGUCGAAUUAAAAAUUGAUUUUCUCUCGAGAGCAUCAACCUUGGUUCAUAAUUACGUAUCUACAGGAGGGCCCA ....((.........((((....)))).....(((((...(((....)))(((((((...(((((.((((....)))).)))))..))))))).................)))))))... ( -25.50) >consensus UUUUGC_GCUUUUCGCUGCAGCAGCAGAUCAAUUUCCAGCGACAGCUGUCGAAUCAAGAAUUGAUUUUCCCUCGAGACCA__AA_________GAAAUUGAGAAACUGCAGGAAA_____ ...............((((((....((((((((((....((((....))))......))))))))))...(((((......................)))))...))))))......... (-15.60 = -17.10 + 1.50)

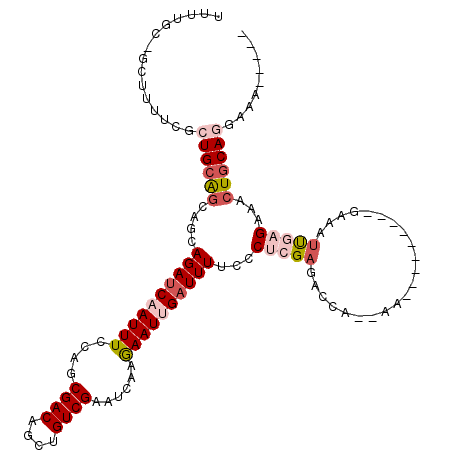
| Location | 16,726,250 – 16,726,354 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -17.68 |
| Energy contribution | -17.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16726250 104 - 23771897 -----UUUCCUGCAGUUUCUCAGUUUC---------UU--GAGUCUCGAGGGAAAAUCAAUUCUUGAUUCGACAGCUGUCGCUGGAAAUUGAUCUGCUGCUGCAGCGAAAAGCUACAAAA -----(((((((((((...((((((((---------((--(((((..(((..........)))..)))))))((((....))))))))))))......))))))).)))).......... ( -32.70) >DroSec_CAF1 33584 90 - 1 -----UUUCCUGCAGUUUCUCAAUUUC---------UU--GGGUAUCGAGGGAAAAUCGAUUCUUGAUUCGACAGCUGUCGCUGGAAAUUGAUCUGCUGCUGCA--------------AA -----......(((((...((((((((---------(.--.((.(((((.......))))).))..).....((((....))))))))))))...)))))....--------------.. ( -28.70) >DroSim_CAF1 38128 90 - 1 -----UUUCUUGCAGUUUCUCAAUUUC---------UU--GGGUCUCGAGGGAAAAUCGAUUCUUGAUUCGACAGCUGUCGCUGGAAAUUGGUCUGCUGCUGCA--------------AA -----....(((((((...((((((((---------(.--.((..((((.......))))..))..).....((((....))))))))))))......))))))--------------). ( -27.30) >DroEre_CAF1 34129 109 - 1 UUUCGCCUCCUGCAGUUUCUUGAUUCC---------UU--UAGUCUCGGGGGAAAAUCAAUUCCUAAUUCGACAGCUGUCGCUGGAAAUUGAUCUGCCGCUGCAGCGAAAAGCAGCAAAA ((((((....((((((..(((((..(.---------..--..)..)))))((...(((((((((.....((((....))))..)).)))))))...)))))))))))))).......... ( -30.10) >DroYak_CAF1 37030 108 - 1 UUUCGCCUCCUGCAGUUUCUCGAUUCC---------UU--UAGUCUCGAGGGAAAAUCAAUUCUUAAUUCGACAGCUGUCGCUGAAAAUUGAUCUGCCGCUGCAGCGAAAAGCGGCAAA- ....((((((((((((..(((((..(.---------..--..)..)))))((...(((((((.(((...((((....)))).))).)))))))...))))))))).)).....)))...- ( -33.30) >DroAna_CAF1 29130 120 - 1 UGGGCCCUCCUGUAGAUACGUAAUUAUGAACCAAGGUUGAUGCUCUCGAGAGAAAAUCAAUUUUUAAUUCGACAACUGUCGCUGGAAAUUGAUCUGCUGGCGCAGUGAAAAGAUGCAAAA ...(((.....(((((........(((.(((....))).)))(((....)))....((((((((((...((((....)))).))))))))))))))).)))(((.(....)..))).... ( -27.30) >consensus _____CCUCCUGCAGUUUCUCAAUUUC_________UU__GAGUCUCGAGGGAAAAUCAAUUCUUAAUUCGACAGCUGUCGCUGGAAAUUGAUCUGCUGCUGCAGCGAAAAGC_GCAAAA .........(((((((...((((((....................(((((((((......)))))...))))((((....))))..))))))......)))))))............... (-17.68 = -17.47 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:34 2006