
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,860,746 – 1,860,838 |
| Length | 92 |
| Max. P | 0.999446 |

| Location | 1,860,746 – 1,860,838 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
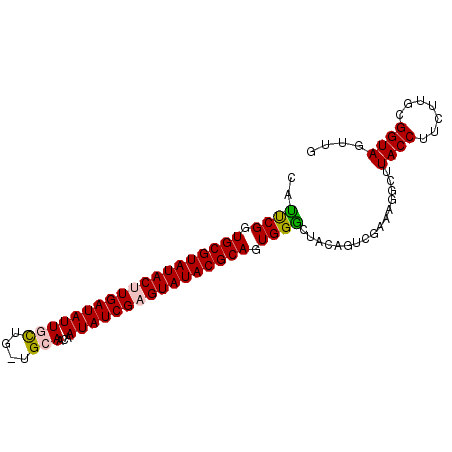
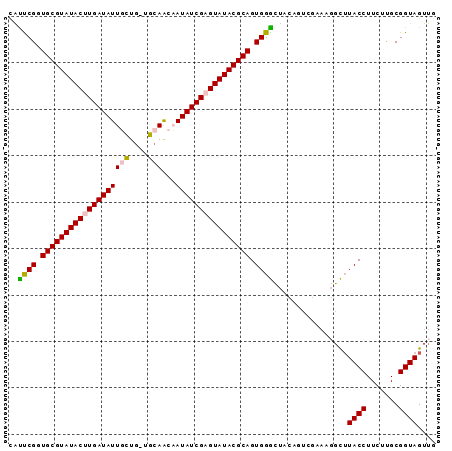
>3L_DroMel_CAF1 1860746 92 + 23771897 CAUUCGAUGCGUAUACUUGAUAUUGCUAUUGCAACAAUAUCGUGUAUACGCAGUGGGUUACAGUCGAAAUGCUUACCUUCUUGCGGUAGUUG ..((((((((((((((.((((((((.........)))))))).))))))))..((.....))))))))....(((((.......)))))... ( -28.00) >DroSec_CAF1 742 91 + 1 UAUUCGGUGCGUAUACUUGAUAUUGCUG-UGCAACCAUAUCGAGUAUACGCAGUGGACUUCAGUCGAAAGGUUUACCUUCUUGCGGUAGUUG ....((.(((((((((((((((((((..-.)))...)))))))))))))))).))((((...(.(((((((....)))).))))...)))). ( -32.80) >DroSim_CAF1 617 91 + 1 AAUUCGGUGCGUAUACUUGAUAUUGCUG-UGCAACCAUAUCGAGUAUACGCAGUGGGCUUCAGUCGGAAGGCUUACCUUCUUGCGGUAGUUG ....((.(((((((((((((((((((..-.)))...)))))))))))))))).))((((...((.((((((....)))))).))...)))). ( -37.40) >DroEre_CAF1 671 77 + 1 UGCUCGGUGCGUAUACUUGAUAUUUUCG-GGCAGCAAUAUCGCGUAUACGCAGUGGG--------------CGUACCUUCUUGCGGUAGUUG .(((((.(((((((((.(((((((....-......))))))).))))))))).))))--------------).((((.......)))).... ( -30.00) >DroYak_CAF1 652 87 + 1 CAUCCGGUGCGUAUACUUGAUAUUUCCG-GGCAACAAUAUCGAGUAUACGCAGUGGGCUACA----AAAGGCUUACCUUCUUGCGGUAGUUG ...(((.(((((((((((((((((....-(....)))))))))))))))))).)))((..((----(((((....)))).)))..))..... ( -35.90) >consensus CAUUCGGUGCGUAUACUUGAUAUUGCUG_UGCAACAAUAUCGAGUAUACGCAGUGGGCUACAGUCGAAAGGCUUACCUUCUUGCGGUAGUUG ..((((.(((((((((((((((((((....)))...)))))))))))))))).))))................((((.......)))).... (-28.32 = -28.48 + 0.16)

| Location | 1,860,746 – 1,860,838 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -23.34 |
| Energy contribution | -24.34 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
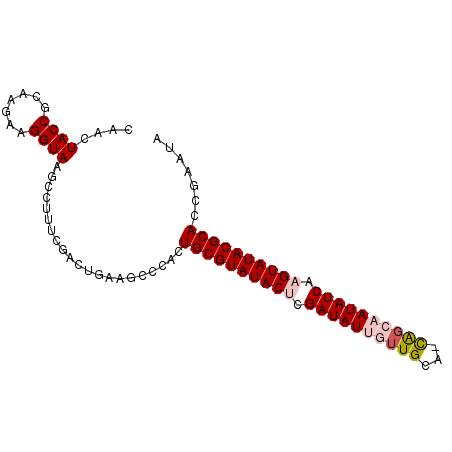
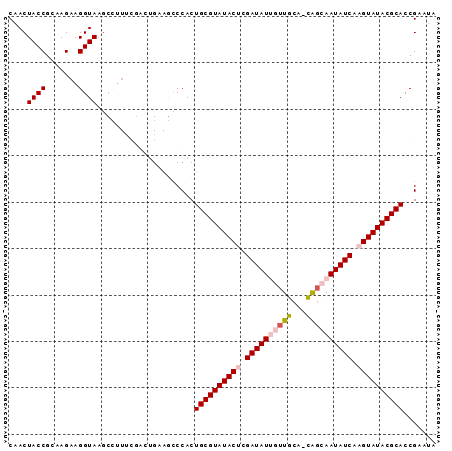
>3L_DroMel_CAF1 1860746 92 - 23771897 CAACUACCGCAAGAAGGUAAGCAUUUCGACUGUAACCCACUGCGUAUACACGAUAUUGUUGCAAUAGCAAUAUCAAGUAUACGCAUCGAAUG ....((((.......)))).....(((((...........(((((((((..((((((((((...))))))))))..)))))))))))))).. ( -27.10) >DroSec_CAF1 742 91 - 1 CAACUACCGCAAGAAGGUAAACCUUUCGACUGAAGUCCACUGCGUAUACUCGAUAUGGUUGCA-CAGCAAUAUCAAGUAUACGCACCGAAUA ....((((.......)))).....((((((....)))...((((((((((.(((((.(((...-.))).))))).))))))))))..))).. ( -26.90) >DroSim_CAF1 617 91 - 1 CAACUACCGCAAGAAGGUAAGCCUUCCGACUGAAGCCCACUGCGUAUACUCGAUAUGGUUGCA-CAGCAAUAUCAAGUAUACGCACCGAAUU ............(((((....)))))..............((((((((((.(((((.(((...-.))).))))).))))))))))....... ( -26.90) >DroEre_CAF1 671 77 - 1 CAACUACCGCAAGAAGGUACG--------------CCCACUGCGUAUACGCGAUAUUGCUGCC-CGAAAAUAUCAAGUAUACGCACCGAGCA ....((((.......)))).(--------------(.(..(((((((((..((((((......-....))))))..)))))))))..).)). ( -23.30) >DroYak_CAF1 652 87 - 1 CAACUACCGCAAGAAGGUAAGCCUUU----UGUAGCCCACUGCGUAUACUCGAUAUUGUUGCC-CGGAAAUAUCAAGUAUACGCACCGGAUG ...((((....((((((....)))))----))))).((..((((((((((.((((((......-....)))))).))))))))))..))... ( -28.50) >consensus CAACUACCGCAAGAAGGUAAGCCUUUCGACUGAAGCCCACUGCGUAUACUCGAUAUUGUUGCA_CAGCAAUAUCAAGUAUACGCACCGAAUA ....((((.......)))).....................((((((((((.((((((((((...)))))))))).))))))))))....... (-23.34 = -24.34 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:09 2006