| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,721,036 – 16,721,146 |
| Length | 110 |
| Max. P | 0.943980 |

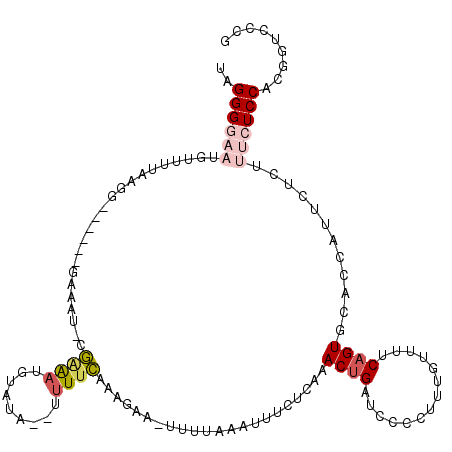
| Location | 16,721,036 – 16,721,146 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -4.66 |
| Energy contribution | -5.78 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16721036 110 + 23771897 UAGGGGAAUAUUUGAAGG------AAAAUUUGAAAUGUAUA--UUUUCAAAGAAC-UUCAAAUUUUUCAAACUGAUCCCCUUUGUUUUCAGUGCACCAUUCUCUUUCUCCACGGUCCCG ..((((((.((((((((.------....(((((((......--.)))))))...)-))))))).......(((((............)))))............)))))).((....)) ( -23.40) >DroSec_CAF1 28460 110 + 1 UAGGGGAAUGUUUGAGGG------GAAAU-CGAGAUGUAUU--UUUUCAAAGAAAUUUUCAAUUUCUCAAACUGUUCCCCUUUGUUUUCAGUGCACCAUUCUCUUUCUCCACGCUCCCC ..(((((.(((..(((((------(((..-.((((......--.))))..((((((.....)))))).......))))))))(((.......)))...............))).))))) ( -25.20) >DroSim_CAF1 32934 110 + 1 UAGGGGAACGUUUUAAGG------GAAAU-CGAGAUGUAUA--UUUUCAAAGAAAUUUUUAAUUUCUCAAACUGAUCCCCUUUGUUUUCAGUGCACCAUUCUCUUUCUCCACGCUCCCC ..(((((.(((...((((------(((..-.((((..((.(--((((....)))))...))...))))..(((((............)))))......))))))).....))).))))) ( -26.00) >DroEre_CAF1 29085 117 + 1 UAGGGAUCUGUUUUAAAAAAAUCAGAAAU-CAGAAUAUACGAGUAUAUAUAUAU-UGUAAAAUGCGUUGAACAGAUCCCUUUUGUUUUCCGUGUACCAUUCUCUUUCUCCACGGUCUGG .((((((((((((........((......-..))...(((.(((((....))))-)))).........))))))))))))........(((((................)))))..... ( -23.49) >DroYak_CAF1 31262 108 + 1 UAGGGAUCUGUUUUAAAG------GAAAU-CGGAAUAUAUA--UUUUCAAUCAU-UUUUAAAUUGGUUGAACUGUUUGCUUUUGUUUUCAGUGCACCAUUCUCUUUCUCCACGGCCC-G ..(((..((((...((((------..(((-.((........--...(((((((.-........)))))))((((...((....))...))))...)))))..))))....)))))))-. ( -23.00) >consensus UAGGGGAAUGUUUUAAGG______GAAAU_CGAAAUGUAUA__UUUUCAAAGAA_UUUUAAAUUUCUCAAACUGAUCCCCUUUGUUUUCAGUGCACCAUUCUCUUUCUCCACGGUCCCG ..((((((.......................((((.........))))......................((((..............))))............))))))......... ( -4.66 = -5.78 + 1.12)


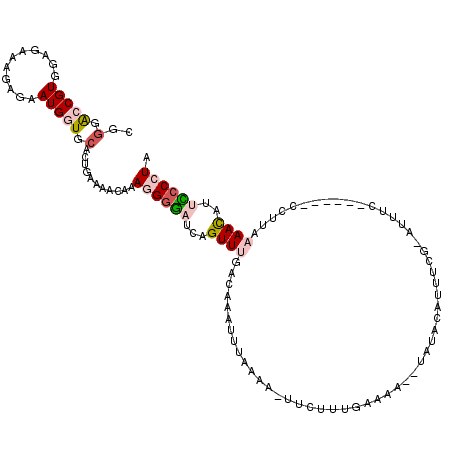
| Location | 16,721,036 – 16,721,146 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -25.79 |
| Consensus MFE | -4.68 |
| Energy contribution | -5.96 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.18 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16721036 110 - 23771897 CGGGACCGUGGAGAAAGAGAAUGGUGCACUGAAAACAAAGGGGAUCAGUUUGAAAAAUUUGAA-GUUCUUUGAAAA--UAUACAUUUCAAAUUUU------CCUUCAAAUAUUCCCCUA ..(.(((((...........))))).)...........((((((...((((((((((((((((-((....(....)--.....))))))))))).------..)))))))..)))))). ( -27.90) >DroSec_CAF1 28460 110 - 1 GGGGAGCGUGGAGAAAGAGAAUGGUGCACUGAAAACAAAGGGGAACAGUUUGAGAAAUUGAAAAUUUCUUUGAAAA--AAUACAUCUCG-AUUUC------CCCUCAAACAUUCCCCUA ((((((.(((((((..((((..((....)).....(((((..(.....((..(....)..))..)..)))))....--......)))).-.))))------)......)).)))))).. ( -27.00) >DroSim_CAF1 32934 110 - 1 GGGGAGCGUGGAGAAAGAGAAUGGUGCACUGAAAACAAAGGGGAUCAGUUUGAGAAAUUAAAAAUUUCUUUGAAAA--UAUACAUCUCG-AUUUC------CCUUAAAACGUUCCCCUA ((((((((((((((..((((.((((.(.((........)).).)))).((.(((((((.....))))))).))...--......)))).-.))))------)......))))))))).. ( -33.40) >DroEre_CAF1 29085 117 - 1 CCAGACCGUGGAGAAAGAGAAUGGUACACGGAAAACAAAAGGGAUCUGUUCAACGCAUUUUACA-AUAUAUAUAUACUCGUAUAUUCUG-AUUUCUGAUUUUUUUAAAACAGAUCCCUA .....(((((................)))))........(((((((((((............((-..((((((......))))))..))-.....(((.....))).))))))))))). ( -25.89) >DroYak_CAF1 31262 108 - 1 C-GGGCCGUGGAGAAAGAGAAUGGUGCACUGAAAACAAAAGCAAACAGUUCAACCAAUUUAAAA-AUGAUUGAAAA--UAUAUAUUCCG-AUUUC------CUUUAAAACAGAUCCCUA .-(((...((...((((..(((((((.((((..............)))).))..(((((.....-..)))))....--........)).-)))..------))))....))...))).. ( -14.74) >consensus CGGGACCGUGGAGAAAGAGAAUGGUGCACUGAAAACAAAGGGGAUCAGUUUGACAAAUUUAAAA_UUCUUUGAAAA__UAUACAUUUCG_AUUUC______CCUUAAAACAUUCCCCUA ..(.(((((...........))))).)...........((((((...((((.......................................................))))..)))))). ( -4.68 = -5.96 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:27 2006