| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
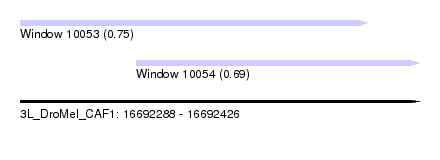
| Location | 16,692,288 – 16,692,426 |
| Length | 138 |
| Max. P | 0.745955 |

| Location | 16,692,288 – 16,692,408 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.33 |
| Mean single sequence MFE | -44.33 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
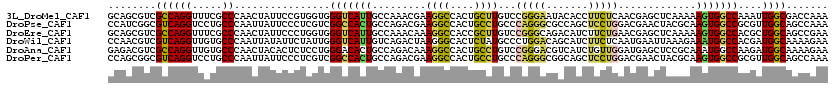
>3L_DroMel_CAF1 16692288 120 + 23771897 GCAGCGUCGCCAGGUUUCGCCCAACUAUUCCGUGGUGGGUCAUUGCCAAACGAAGGCCACUGCUUGUCCGGGAAUACACCUUCUCAACGAGCUCAAAAAGUGGCCAAAUUGGCGACCAAA ...(.((((((((((...(((((.((((...)))))))))....))).......(((((((((((((..(((((......))))).))))))......)))))))....))))))))... ( -48.40) >DroPse_CAF1 13493 120 + 1 CCAUCGGCGUCAGGUCCUGCCCAAUUAUUCCCUCGUCGGCCACUGCCAGACGAAGGCCACUGCCUGCCCAGGGCGCCAGCUCCUGGACGAACUACGCAAGUGGCCGCGUUGGCAGCCAAA .....((((((((....(((............((((((((....))).))))).(((((((((.((.(((((((....)).))))).))......)).))))))))))))))).)))... ( -46.90) >DroEre_CAF1 14169 120 + 1 GCAGCGUCGCCAGGUUUCGCCCAACUAUUCCCUGGUGGGUCAUUGCCAAACAAAGGCCACCGCUUGUCCGGGCAGACAUCUUCUGAACGAGCUCAAAAAGUGGCCACGCUGGCAGCCGAA .(((((((((((((................)))))))((((((((((.......)))....((((((.((((.((....)))))).))))))......)))))))))))))......... ( -43.09) >DroWil_CAF1 34513 120 + 1 CCAACGUCGUCAGGUUGUGCCCAAUUAUAUUCUAUUGGGUCAUUGUCAGACUAAGGGCACUCUAUGCCCUGGACAGCAUCUUCUCAAUGAAUUAAAGAAAUGGCCACGAUGGCAAAAGAA .....((((((.(((..(.((((((........))))))((((((..(((((.((((((.....))))))....))..)))...)))))).........)..)))..))))))....... ( -35.10) >DroAna_CAF1 11140 120 + 1 GAGACGUCGCCAGGUUGUGCCCAACUACACUCUCCUGGGACACUGCCAGACAAAGGCCACUGCCUGUCCGGGACGUCAUCUGUUGGAUGAGCUCCGCAAAUGGCCAAGAUGGCAAAAGAA .....(((.(((((..(((........)))...))))))))..(((((......(((((.((((.....)(((..(((((.....)))))..))))))..)))))....)))))...... ( -40.90) >DroPer_CAF1 13375 120 + 1 CCAGCGGCGUCAGGUCCUGCCCAAUUAUUCCCUCGUCGGCCACUGCCAGACGAAGGCCACUGCCUGCCCAGGGCGGCAGCUCCUGGACGAACUACGCAAGUGGCCGCGUUGGCAGCCAAA ...(((((((((((..(((((........((.((((((((....))).))))).)).....((((.....)))))))))..))).)))....(((....)))))))).((((...)))). ( -51.60) >consensus CCAGCGUCGCCAGGUUCUGCCCAACUAUUCCCUCGUGGGUCACUGCCAGACGAAGGCCACUGCCUGCCCGGGACGACAUCUUCUGAACGAACUAAACAAGUGGCCACGUUGGCAACCAAA ........((((((.....))................(((((((.........((((....))))...(((((.......))))).............)))))))....))))....... (-19.68 = -19.10 + -0.58)

| Location | 16,692,328 – 16,692,426 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -22.86 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16692328 98 + 23771897 CAUUGCCAAACGAAGGCCACUGCUUGUCCGGGAAUACACCUUCUCAACGAGCUCAAAAAGUGGCCAAAUUGGCGACCAAAGCCUUAGGUUUCCAUAUG ..(((((((.....(((((((((((((..(((((......))))).))))))......)))))))...)))))))...(((((...)))))....... ( -31.40) >DroSec_CAF1 13905 98 + 1 CAUUGCCAAACGAAGGCCACAGCGUGUCCGGGAAGACAUCUUCUGAACGAGCUCAAGAAGUGGCCACGUUGGCAGCCAAAGCCUUAGGUUCCCAUAUG ..(((((((.((..(((((((((.(((.(((.(((....)))))).))).)))......)))))).)))))))))....((((...))))........ ( -29.70) >DroSim_CAF1 21802 98 + 1 CAUUGCCAAACGAAGGCCACAGCUUGUCCGGGAAGACAUCUUCUGAACGAGCUCAAGAAGUGGCCACGUUGGCAGCCAAAGCCUUAGGUUUCCAUAUG ..(((((((.((..(((((((((((((.(((.(((....)))))).)))))))......)))))).)))))))))...(((((...)))))....... ( -35.10) >DroEre_CAF1 14209 98 + 1 CAUUGCCAAACAAAGGCCACCGCUUGUCCGGGCAGACAUCUUCUGAACGAGCUCAAAAAGUGGCCACGCUGGCAGCCGAAGCCUUAAGUUCCCAUAAG ..((((((......((((((.((((((.((((.((....)))))).)))))).......))))))....))))))....................... ( -27.00) >DroYak_CAF1 16544 98 + 1 CAUUGCCAAACGAAGGCCACAGCUUGUCCGGGAAGACAUCUUCUGAACGAGCUCAAAAAGUGGCCACGCUGGCAGCCAAAGCCUUAAGUUCCCAUAGG ..((((((..((..(((((((((((((.(((.(((....)))))).)))))))......)))))).)).))))))....................... ( -31.20) >DroAna_CAF1 11180 87 + 1 CACUGCCAGACAAAGGCCACUGCCUGUCCGGGACGUCAUCUGUUGGAUGAGCUCCGCAAAUGGCCAAGAUGGCAAAAGAAGCCAUAG----------- ....((((((((..(((....)))))))(((..(.(((((.....))))))..)))....))))....(((((.......)))))..----------- ( -28.40) >consensus CAUUGCCAAACGAAGGCCACAGCUUGUCCGGGAAGACAUCUUCUGAACGAGCUCAAAAAGUGGCCACGUUGGCAGCCAAAGCCUUAGGUUCCCAUAUG ..((((((......((((((.((((((.(((((.......))))).)))))).......))))))....))))))....................... (-22.86 = -23.50 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:16 2006