| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,681,098 – 16,681,218 |
| Length | 120 |
| Max. P | 0.710549 |

| Location | 16,681,098 – 16,681,218 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -44.31 |
| Consensus MFE | -37.65 |
| Energy contribution | -37.07 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
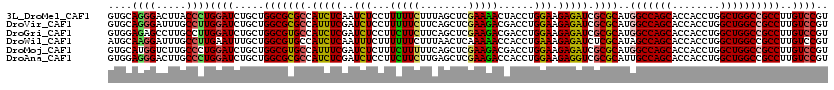
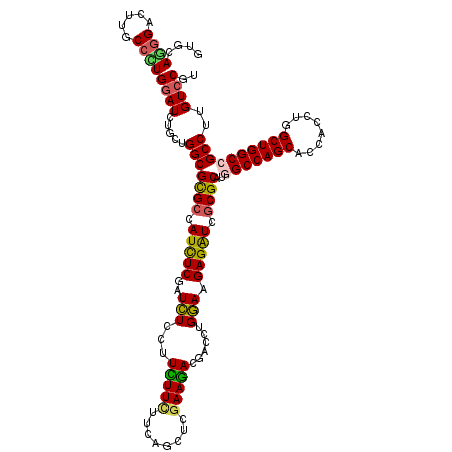
>3L_DroMel_CAF1 16681098 120 - 23771897 GUGCAGGGACUUACCCUGGAUCUGCUGGCGCGCCAUCUCAAUCUCCUUUUUCUUUAGCUCGAAAACUACCUGGAAGAGAUCGCGCAUGGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU ....((((.....))))((((.....(((((((.(((((....(((.(((((........)))))......))).))))).))))..(((((((........))))))))))..)))).. ( -45.00) >DroVir_CAF1 548 120 - 1 GUGCAGGGAUUUGCCUUGGAUCUGCUGGCGCGCCAUUUCGAUCUCCUUUUUCUUCAGCUCGAAGACGACCUGGAAGAGAUCGCGCAUGGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU (..(((((....(((..((...(((((((((((.(((((....(((.((.(((((.....))))).))...))).))))).))))...)))))))...)).)))......)))))..).. ( -46.20) >DroGri_CAF1 1513 120 - 1 GUGGAGAGCCUUGCCUUGGAUCUGCUGGCGUGCCAUCUCGAUCUCCUUCUUCUUCAGCUCGAAGACGACCUGGAAGAGAUCGCGCAUGGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU ..((.(.(((..(((..((...(((((((((((.(((((....(((.((.(((((.....))))).))...))).))))).))))...)))))))...)).))).)))).))........ ( -48.20) >DroWil_CAF1 21329 120 - 1 AUGCAAGGAUUUGCCUUGAAUUUGCUGGCGUGCCAUCUCAAUUUCUUUUUUCUUUAACUCAAAAACCACCUGAAAGAGAUCUCGCAUAGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU ..((((((....(((.......(((((((((((.(((((..((((..........................)))))))))...)))).)))))))((....))..)))..)))))).... ( -33.57) >DroMoj_CAF1 551 120 - 1 GUGCAUGGUCUUGCCCUGGAUCUGCUGGCGUGCCAUUUCGAUCUCUUUCUUUUUCAGCUCGAAGACGACCUGGAAGAGAUCGCGCAUGGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU (..(((((((..(((..((...(((((((((((.....((((((((......(((((.(((....))).))))))))))))).)))).)))))))...)).))).)))))...))..).. ( -47.60) >DroAna_CAF1 337 120 - 1 GUGGAGGGACUUGCCCUGGAUCUGCUGGCGCGCCAUCUCGAUCUCCUUCUUCUUGAGCUCGAAGACCACCUGGAAGAGGUCGCGCAUUGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU ..((((((.((.(((..((...(((((((((((.(((((........(((((........)))))((....))..))))).))))...)))))))...)).))).))...)))..))).. ( -45.30) >consensus GUGCAGGGACUUGCCCUGGAUCUGCUGGCGCGCCAUCUCGAUCUCCUUCUUCUUCAGCUCGAAGACGACCUGGAAGAGAUCGCGCAUGGCCAGCACCACCUGGCUGGCCGCCUUGUCCGU ....((((.....))))((((.....(((((((.(((((..(((...(((((........)))))......))).))))).))))..(((((((........))))))))))..)))).. (-37.65 = -37.07 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:07 2006