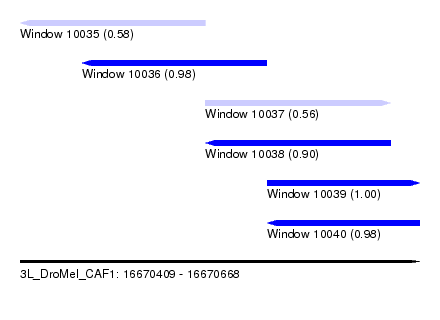
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,670,409 – 16,670,668 |
| Length | 259 |
| Max. P | 0.996177 |

| Location | 16,670,409 – 16,670,529 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -33.60 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16670409 120 - 23771897 CCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCUGCUGCUCCAUCAAUGGAAUUUCGGCGAUUCCGGCGCUUCACCACCACCGUCGUGGAUCUCGAUUUUGGACCCGCCAGUUUAGUAUUA ........((((((((((((((((((((........)))))))...))).....))))))))))((((..((....((((....))))..(........)))..))))............ ( -37.60) >DroSec_CAF1 38025 120 - 1 CCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCGGCGAUUCCGGCGCUUCACCACCACCGCCGUGGCUCUCGAUUUUGGACCCGCCAGUUUAGUAUUA ........((((((((.....(((((((........)))))))....))))))))((((..(((((.......)).((((....))))...........)))..))))............ ( -36.20) >DroSim_CAF1 38103 120 - 1 CCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCGGCGAUUCCGGCGCUUCACCACCACCGCCGUGGAUUUCGAUUUUGGACCCGCCAGUUUAGUAUUA ........((((((((.....(((((((........)))))))....))))))))((((..(((((.......)).((((....))))...........)))..))))............ ( -36.90) >DroEre_CAF1 38113 120 - 1 CCCCCGUCGGAAUUGCCGCCUGUGGAGCCUUCGGCUGCUCCAUUAAUGCGAUUUCGGCGAUUUCGGCGCUUCACCACCACCGUAGUGGAUCUCGAUUUUGGACCCGCCAGUUUACUAUUC ....((((((((((((.....((((((((....)..)))))))....)))))))))))).....((((..((....((((....))))..(........)))..))))............ ( -40.70) >DroYak_CAF1 39300 120 - 1 CCCACGUCGGCAUUGCCGCCUGUGGCGCCUCCGGCUGCUCCAUCAGUGCGAUUUCGGCGAUUCCGGCGCUUCACCACCACCGUAGUGGAUCUCGAUCUUGGACCCGCCAGUUUACUAUUC .(((((((((.((((((((((((((.((........)).))).))).))(....))))))).))))))........((((....))))..........)))................... ( -36.80) >consensus CCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCGGCGAUUCCGGCGCUUCACCACCACCGUCGUGGAUCUCGAUUUUGGACCCGCCAGUUUAGUAUUA ........((((((((.....(((((((........)))))))....))))))))((((..(((((.......)).((((....))))...........)))..))))............ (-33.60 = -33.80 + 0.20)


| Location | 16,670,449 – 16,670,569 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -40.86 |
| Consensus MFE | -31.10 |
| Energy contribution | -32.14 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16670449 120 - 23771897 UGCCGAAUAACCACUUCCGGAGAAAGCUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCUGCUGCUCCAUCAAUGGAAUUUCGGCGAUUCCGGCGCUUCACCACCAC .(((..........((((((((.......))))))))...........((((((((((((((((((((........)))))))...))).....)))))))))))))............. ( -42.30) >DroSec_CAF1 38065 120 - 1 UGUCGAAUUACCACUUCCGGAGAAAACUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCGGCGAUUCCGGCGCUUCACCACCAC .(((((((..((..((((((((.......))))))))...........((((((((.....(((((((........)))))))....))))))))))..))).))))............. ( -40.00) >DroSim_CAF1 38143 120 - 1 UGUCGAAUUACCACUUCCGGAGAAAGCUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCGGCGAUUCCGGCGCUUCACCACCAC .(((((((..((..((((((((.......))))))))...........((((((((.....(((((((........)))))))....))))))))))..))).))))............. ( -40.00) >DroEre_CAF1 38153 120 - 1 UCUUGAAAUACCGCCUCCGCAGAAACCUGCUCUGGAUUUACCCCCGUCGGAAUUGCCGCCUGUGGAGCCUUCGGCUGCUCCAUUAAUGCGAUUUCGGCGAUUUCGGCGCUUCACCACCAC ...((((....(((((((((((....))))...)))........((((((((((((.....((((((((....)..)))))))....)))))))))))).....)))).))))....... ( -43.10) >DroYak_CAF1 39340 120 - 1 UCUGGAAUUUCCACCUCCGCAGAAACCUGCGCUGCAUUUACCCACGUCGGCAUUGCCGCCUGUGGCGCCUCCGGCUGCUCCAUCAGUGCGAUUUCGGCGAUUCCGGCGCUUCACCACCAC ..(((............(((((....))))).............((((((.((((((((((((((.((........)).))).))).))(....))))))).)))))).....))).... ( -38.90) >consensus UGUCGAAUUACCACUUCCGGAGAAACCUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCGGCGAUUCCGGCGCUUCACCACCAC ....(((...((...(((((((.......)))))))............((((((((((...(((((((........)))))))...........))))))))))))...)))........ (-31.10 = -32.14 + 1.04)



| Location | 16,670,529 – 16,670,649 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.04 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16670529 120 + 23771897 UAAUUCCAGAGCAGCUUUCUCCGGAAGUGGUUAUUCGGCAGGAAAUAAUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUUCCGAGCAAG ...((((.(((.......))).))))...........((.((((((((((((...(((..((((((((.(((((....)))))))))).)))..)))...)))).))))))))..))... ( -43.50) >DroSec_CAF1 38145 120 + 1 UAAUUCCAGAGCAGUUUUCUCCGGAAGUGGUAAUUCGACAGGAAAUAUUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGG ...(((..(((.((.((((((((((((((....(((....)))..))))))))..(((..((((((((.(((((....)))))))))).)))..))).)))))).)).)))..))).... ( -37.80) >DroSim_CAF1 38223 120 + 1 UAAUUCCAGAGCAGCUUUCUCCGGAAGUGGUAAUUCGACAGGAAAUAUUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACCUAUUCCCGAGCAGG ..........((.(((((((.((((((((....(((....)))..)))))))).)))))))...(((..(((((....)))))((((........)))).((((....)))))))))... ( -36.40) >DroEre_CAF1 38233 120 + 1 UAAAUCCAGAGCAGGUUUCUGCGGAGGCGGUAUUUCAAGAGGAAAUAAUUCCGAAGGAGGUAAAUCAACUGGAUGCAAUUCCGGUGGAAGUAAUUCCAAAGGAACUAAUUCCCGGACAGG ...((((((.((((....)))).((.....((((((....((((....))))....))))))..))..)))))).....((((.(((((....)))))..((((....)))))))).... ( -35.80) >DroYak_CAF1 39420 120 + 1 UAAAUGCAGCGCAGGUUUCUGCGGAGGUGGAAAUUCCAGAGGAAAUAAUUCCGAAGGAGGUAAACCGACUGGAUGCAAUUCCGGUGGAUGUAAUUCCAGAGGAAUUAAUUCCCGAGCAGG ....(((..(((((....)))))....(((.....)))(.((((.(((((((...((((.((..((.((((((......))))))))...)).))))...))))))).)))))..))).. ( -40.50) >consensus UAAUUCCAGAGCAGCUUUCUCCGGAAGUGGUAAUUCGACAGGAAAUAAUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGG ..................(((.((((..(((..(((....((((....))))...(((..((..((((.(((((....)))))))))...))..))).)))..)))..)))).))).... (-22.68 = -23.04 + 0.36)



| Location | 16,670,529 – 16,670,649 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -23.21 |
| Energy contribution | -24.81 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16670529 120 - 23771897 CUUGCUCGGAAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUGUACCUCCUUCGGAAUUAUUUCCUGCCGAAUAACCACUUCCGGAGAAAGCUGCUCUGGAAUUA ...((..((((((((.((((...(((..(((......(((((....)))))......)))..)))...)))))))))))).))...........((((((((.......))))))))... ( -37.30) >DroSec_CAF1 38145 120 - 1 CCUGCUCGGGAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUGUACCUCCUUCGGAAAUAUUUCCUGUCGAAUUACCACUUCCGGAGAAAACUGCUCUGGAAUUA ......((((((....((((...(((..(((......(((((....)))))......)))..)))...))))....))))))............((((((((.......))))))))... ( -35.60) >DroSim_CAF1 38223 120 - 1 CCUGCUCGGGAAUAGGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUUUACCUCCUUCGGAAAUAUUUCCUGUCGAAUUACCACUUCCGGAGAAAGCUGCUCUGGAAUUA .......(((((((..((((...(((...........(((((....)))))...........)))...)))).)))))))..............((((((((.......))))))))... ( -32.35) >DroEre_CAF1 38233 120 - 1 CCUGUCCGGGAAUUAGUUCCUUUGGAAUUACUUCCACCGGAAUUGCAUCCAGUUGAUUUACCUCCUUCGGAAUUAUUUCCUCUUGAAAUACCGCCUCCGCAGAAACCUGCUCUGGAUUUA ...(((((((((.(((((((...(((..((..((.((.(((......))).)).))..))..)))...))))))).))))).................((((....))))...))))... ( -32.00) >DroYak_CAF1 39420 120 - 1 CCUGCUCGGGAAUUAAUUCCUCUGGAAUUACAUCCACCGGAAUUGCAUCCAGUCGGUUUACCUCCUUCGGAAUUAUUUCCUCUGGAAUUUCCACCUCCGCAGAAACCUGCGCUGCAUUUA ..(((..(((((((((((((..((((......))))..)))))))..(((((..((....))......((((....)))).))))))))))).....(((((....)))))..))).... ( -36.30) >consensus CCUGCUCGGGAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUUUACCUCCUUCGGAAUUAUUUCCUGUCGAAUUACCACUUCCGGAGAAACCUGCUCUGGAAUUA .......((((((.((((((...(((...........(((((....)))))...........)))...))))))))))))...............(((((((.......))))))).... (-23.21 = -24.81 + 1.60)



| Location | 16,670,569 – 16,670,668 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16670569 99 + 23771897 GGAAAUAAUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUUCCGAGCAAGUUAUA---------------------CCGGUGCUGGUAGU ((((((((((((...(((..((((((((.(((((....)))))))))).)))..)))...)))).))))))))..........((---------------------(((....))))).. ( -39.50) >DroSec_CAF1 38185 99 + 1 GGAAAUAUUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGGUUAUA---------------------CCGGUGCUGGCAGU ((((....((((...(((..((((((((.(((((....)))))))))).)))..)))...))))....))))(.((((((.....---------------------))..)))).).... ( -34.70) >DroSim_CAF1 38263 99 + 1 GGAAAUAUUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACCUAUUCCCGAGCAGGUUAUA---------------------CCGGUGCUGGCAGU ((((....((((...(((..((.(((((.(((((....))))))))))..))..)))...))))....))))(.((((((.....---------------------))..)))).).... ( -31.10) >DroEre_CAF1 38273 120 + 1 GGAAAUAAUUCCGAAGGAGGUAAAUCAACUGGAUGCAAUUCCGGUGGAAGUAAUUCCAAAGGAACUAAUUCCCGGACAGGUUAUACCGGUGGAUGUAAUGGUAAUGCCGGUGCUGGUAGU ((((.((.((((...((((.((..((.((((((......)))))).))..)).))))...)))).)).))))(((((.(((..(((((..........)))))..))).)).)))..... ( -34.80) >DroYak_CAF1 39460 120 + 1 GGAAAUAAUUCCGAAGGAGGUAAACCGACUGGAUGCAAUUCCGGUGGAUGUAAUUCCAGAGGAAUUAAUUCCCGAGCAGGUAAAACCGGUGGAGGUAAUGGUAAUACCGGUGCUGGUAGU ((((.(((((((...((((.((..((.((((((......))))))))...)).))))...))))))).))))............(((((((..((((.......))))..)))))))... ( -40.00) >consensus GGAAAUAAUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGGUUAUA_____________________CCGGUGCUGGUAGU ((((....((((...(((..((..((((.(((((....)))))))))...))..)))...))))....))))(.((((((..........................))..)))).).... (-20.33 = -20.65 + 0.32)



| Location | 16,670,569 – 16,670,668 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.49 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16670569 99 - 23771897 ACUACCAGCACCGG---------------------UAUAACUUGCUCGGAAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUGUACCUCCUUCGGAAUUAUUUCC ..((((......))---------------------))..........((((((((.((((...(((..(((......(((((....)))))......)))..)))...)))))))))))) ( -28.00) >DroSec_CAF1 38185 99 - 1 ACUGCCAGCACCGG---------------------UAUAACCUGCUCGGGAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUGUACCUCCUUCGGAAAUAUUUCC ......((((..((---------------------.....)))))).(((((((..((((...(((..(((......(((((....)))))......)))..)))...)))).))))))) ( -26.90) >DroSim_CAF1 38263 99 - 1 ACUGCCAGCACCGG---------------------UAUAACCUGCUCGGGAAUAGGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUUUACCUCCUUCGGAAAUAUUUCC ......((((..((---------------------.....)))))).(((((((..((((...(((...........(((((....)))))...........)))...)))).))))))) ( -24.75) >DroEre_CAF1 38273 120 - 1 ACUACCAGCACCGGCAUUACCAUUACAUCCACCGGUAUAACCUGUCCGGGAAUUAGUUCCUUUGGAAUUACUUCCACCGGAAUUGCAUCCAGUUGAUUUACCUCCUUCGGAAUUAUUUCC ..........((((((.((((............)))).....)).))))(((.(((((((...(((..((..((.((.(((......))).)).))..))..)))...))))))).))). ( -27.90) >DroYak_CAF1 39460 120 - 1 ACUACCAGCACCGGUAUUACCAUUACCUCCACCGGUUUUACCUGCUCGGGAAUUAAUUCCUCUGGAAUUACAUCCACCGGAAUUGCAUCCAGUCGGUUUACCUCCUUCGGAAUUAUUUCC ....((((((..((((..(((............)))..)))))))).))(((.(((((((...(((..((.(((.((.(((......))).)).))).))..)))...))))))).))). ( -32.70) >consensus ACUACCAGCACCGG_____________________UAUAACCUGCUCGGGAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUUUACCUCCUUCGGAAUUAUUUCC ...............................................((((((.((((((...(((...........(((((....)))))...........)))...)))))))))))) (-16.69 = -17.49 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:02 2006