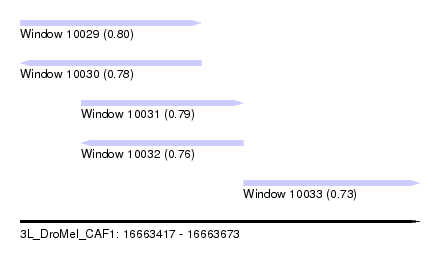
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,663,417 – 16,663,673 |
| Length | 256 |
| Max. P | 0.799970 |

| Location | 16,663,417 – 16,663,533 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -22.85 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16663417 116 + 23771897 UUUU-AGUCCAGCAUGGACCACUGUUGGUUGGCCAAGUGGAAUCGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCUGU-GGAGCACUCUUCACCGCC-CCU ....-...((((((.(.....)))))))..(((...(((((...(((((((((((.((((((..(....)..)))))).)))).)))..((((...-.))))))))))))).)))-... ( -39.70) >DroSec_CAF1 31207 111 + 1 UUUU-UGUCCAGCAUGGACCACUGUU----GCCCAAGUGGAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU-GGAGUCCACUUCACCCC--GCU ...(-((..(((((.(.....)))))----)..)))((((..(((((((((((((.((((((..(....)..)))))).)))).((((.(....))-))).)))))))))..))--)). ( -41.80) >DroSim_CAF1 31263 111 + 1 UUUU-UGUCCAGCAUGGACCACUGUU----GCCCAAGUGGAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU-GGAGUCCACUUCACCCC--CCU ....-..((((((.....(((((...----.....)))))....(((((((((((.((((((..(....)..)))))).)))).)))..)))))))-)))..............--... ( -38.50) >DroEre_CAF1 31336 110 + 1 UUUU-AGACCAGCAUGGACCACUGUU----GCCGAAGUGGAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCUGAGCUCGGC-GGAACCCUCUUCUCCGC--CC- .(((-((((((.(((...(((((...----.....))))).)))...))).((((.((((((..(....)..)))))).)))).))))))...(((-(((.........)))))--).- ( -36.90) >DroYak_CAF1 32421 110 + 1 UUUU-AGUCCAGCAUGGACCACUGUU----GCCCAAGUGGAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGA-GAAACCCUCUUCUCCGC--CC- ....-.....(((.(((((((((...----..((....)).....))))).((((.((((((..(....)..)))))).)))).)))).)))((((-(((......))))))).--..- ( -37.40) >DroAna_CAF1 29561 105 + 1 UUCUAUGUCUUCCUCAGACCACUGUA----CUCCAAGUG----------GAAUGGAGUUGUCCCCACCAGAAGACAACACUGUUUCUGUUCUCAGUUCCAGUACCAGUACCAGCCCCAU .....(((((((....(((.(((.((----.(((....)----------)).)).))).))).......)))))))..((((...(((....)))...))))................. ( -23.70) >consensus UUUU_AGUCCAGCAUGGACCACUGUU____GCCCAAGUGGAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGU_GGAGCCCUCUUCACCGC__CCU ..........(((.(((((((((............)))))...........((((.((((((..(....)..)))))).)))).)))).)))........................... (-22.85 = -22.52 + -0.33)

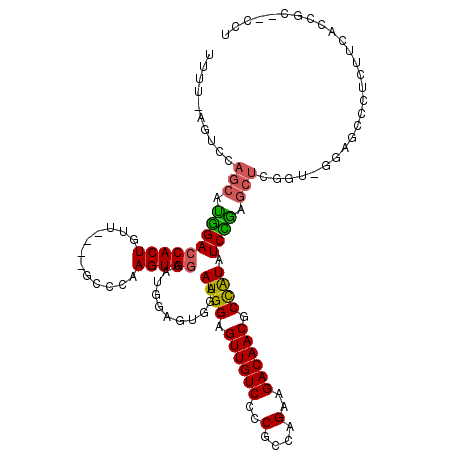
| Location | 16,663,417 – 16,663,533 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16663417 116 - 23771897 AGG-GGCGGUGAAGAGUGCUCC-ACAGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCGAUUCCACUUGGCCAACCAACAGUGGUCCAUGCUGGACU-AAAA .((-(((((((..((((((((.-...))))..(((.((((.(((((((((....))))))))).))))))))))).....))))..)))..)).....((((((.....)))))-)... ( -45.40) >DroSec_CAF1 31207 111 - 1 AGC--GGGGUGAAGUGGACUCC-AGCGAGCUUGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUUCCACUUGGGC----AACAGUGGUCCAUGCUGGACA-AAAA (((--((((((.((((((.(((-((.....))))).((((.(((((((((....))))))))).)))))))))).))))))...(((((----.......))))).))).....-.... ( -44.60) >DroSim_CAF1 31263 111 - 1 AGG--GGGGUGAAGUGGACUCC-AGCGAGCUUGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUUCCACUUGGGC----AACAGUGGUCCAUGCUGGACA-AAAA (((--((((((.((((((.(((-((.....))))).((((.(((((((((....))))))))).)))))))))).)))))).))).(..----..)....((((.....)))).-.... ( -43.80) >DroEre_CAF1 31336 110 - 1 -GG--GCGGAGAAGAGGGUUCC-GCCGAGCUCAGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUUCAUUCCACUUCGGC----AACAGUGGUCCAUGCUGGUCU-AAAA -.(--(((((((((.((((((.-...)))))).(..((((.(((((((((....))))))))).))))..).))))...(((((..(..----..)))))))))..))).....-.... ( -39.00) >DroYak_CAF1 32421 110 - 1 -GG--GCGGAGAAGAGGGUUUC-UCCGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUUCAUUCCACUUGGGC----AACAGUGGUCCAUGCUGGACU-AAAA -((--(((((((((....))))-)))..))))(((.((((.(((((((((....))))))))).))))))).......(((((.(((((----.......))))).).))))..-.... ( -43.50) >DroAna_CAF1 29561 105 - 1 AUGGGGCUGGUACUGGUACUGGAACUGAGAACAGAAACAGUGUUGUCUUCUGGUGGGGACAACUCCAUUC----------CACUUGGAG----UACAGUGGUCUGAGGAAGACAUAGAA ..((..(..(((....)))..)..))...........((.((((((((((....)))))))((((((...----------....)))))----)))).))((((.....))))...... ( -31.50) >consensus AGG__GCGGUGAAGAGGACUCC_ACCGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUUCCACUUGGGC____AACAGUGGUCCAUGCUGGACA_AAAA ................................(((.((((.(((((((((....))))))))).))))))).........((((............))))((((.....))))...... (-23.34 = -23.90 + 0.56)

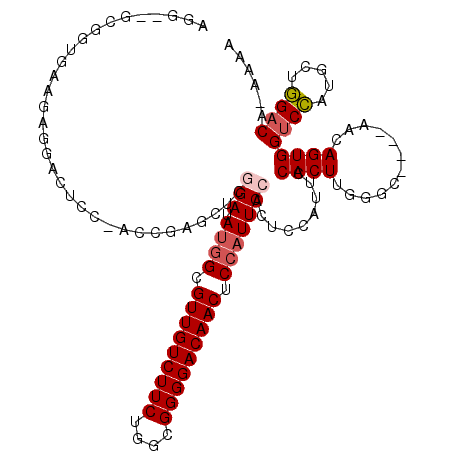
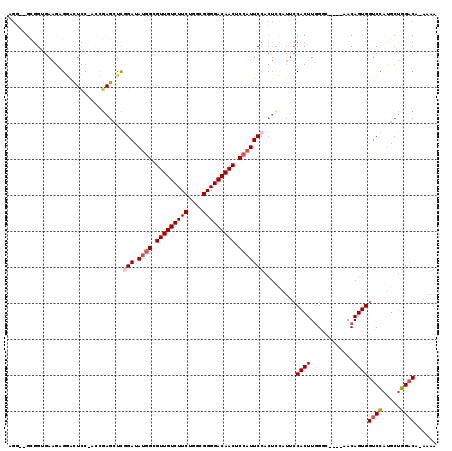
| Location | 16,663,456 – 16,663,560 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -15.34 |
| Energy contribution | -16.62 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16663456 104 + 23771897 AAUCGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCUGU-GGAGCACUCUUCACCGCC-CCUUCUUCCCCCCUUCAC-UUAGCGCACUCG ...((((((((((((.((((((..(....)..)))))).)))).)))(.(((..((-((((..............-............))))))-..))).)))))) ( -30.67) >DroSec_CAF1 31242 95 + 1 AAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU-GGAGUCCACUUCACCCC--GCUU--------CUUCAC-UUAGCGCACUCG ..(((((((((((((.((((((..(....)..)))))).)))).((((.(....))-))).)))))))))...(--(((.--------......-..))))...... ( -32.20) >DroSim_CAF1 31298 95 + 1 AAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU-GGAGUCCACUUCACCCC--CCUU--------CUUCAC-UUAGCGCACUCG ..(((((((((((((.((((((..(....)..)))))).)))).((((.(....))-))).)))))))))....--....--------......-............ ( -29.20) >DroEre_CAF1 31371 94 + 1 AAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCUGAGCUCGGC-GGAACCCUCUUCUCCGC--CC-C--------CUUCAC-UUAGAGCACUCC ..(((((.((.((((.((((((..(....)..)))))).))))..........(((-(((.........)))))--))-)--------))))).-............ ( -29.90) >DroYak_CAF1 32456 94 + 1 AAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGA-GAAACCCUCUUCUCCGC--CC-C--------CUUCAC-UUAGAGCACUCG ..(((((((((((((.((((((..(....)..)))))).)))).))))....((((-(((......))))))).--..-.--------))))).-............ ( -29.90) >DroAna_CAF1 29596 93 + 1 ---------GAAUGGAGUUGUCCCCACCAGAAGACAACACUGUUUCUGUUCUCAGUUCCAGUACCAGUACCAGCCCCAUU-----CCUCUUCAGUUUGGAGCACUCG ---------((((((.((((((..(....)..))))))((((...(((....)))...)))).............)))))-----)((((.......))))...... ( -21.90) >consensus AAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGU_GGAGCCCUCUUCACCGC__CCUU________CUUCAC_UUAGAGCACUCG ..(((((((((((((.((((((..(....)..)))))).)))).)))).((((.....))))..........................))))).............. (-15.34 = -16.62 + 1.28)

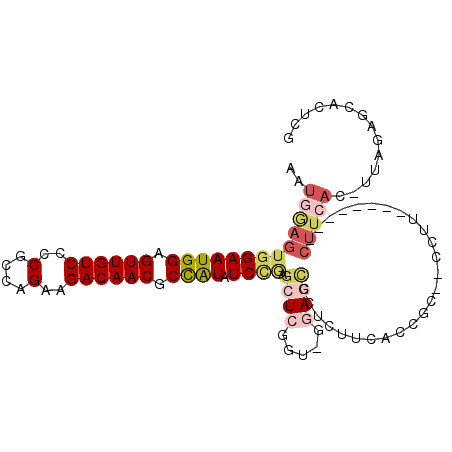
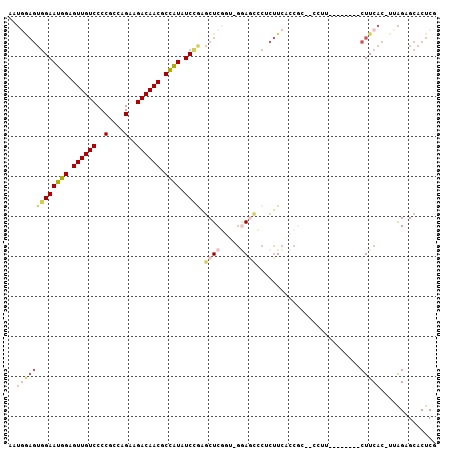
| Location | 16,663,456 – 16,663,560 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16663456 104 - 23771897 CGAGUGCGCUAA-GUGAAGGGGGGAAGAAGG-GGCGGUGAAGAGUGCUCC-ACAGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCGAUU (((((.(.((..-....)).)(((.....((-((((.(....).))))))-......)))(((.((((.(((((((((....))))))))).))))))))))))... ( -37.60) >DroSec_CAF1 31242 95 - 1 CGAGUGCGCUAA-GUGAAG--------AAGC--GGGGUGAAGUGGACUCC-AGCGAGCUUGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUU .((((.((((..-((....--------..))--(((((.......)))))-)))).))))(((.((((.(((((((((....))))))))).)))))))........ ( -35.50) >DroSim_CAF1 31298 95 - 1 CGAGUGCGCUAA-GUGAAG--------AAGG--GGGGUGAAGUGGACUCC-AGCGAGCUUGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUU .(((((..((((-((...(--------....--(((((.......)))))-..)..))))))..((((.(((((((((....))))))))).))))..))))).... ( -34.20) >DroEre_CAF1 31371 94 - 1 GGAGUGCUCUAA-GUGAAG--------G-GG--GCGGAGAAGAGGGUUCC-GCCGAGCUCAGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUUCAUU ((((((((((..-....))--------)-)(--((((((.......))))-))).......((.((((.(((((((((....))))))))).))))))))))))... ( -37.10) >DroYak_CAF1 32456 94 - 1 CGAGUGCUCUAA-GUGAAG--------G-GG--GCGGAGAAGAGGGUUUC-UCCGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUUCAUU ...........(-((((((--------(-((--.((((((((....))))-))))..)))(((.((((.(((((((((....))))))))).))))))).))))))) ( -40.80) >DroAna_CAF1 29596 93 - 1 CGAGUGCUCCAAACUGAAGAGG-----AAUGGGGCUGGUACUGGUACUGGAACUGAGAACAGAAACAGUGUUGUCUUCUGGUGGGGACAACUCCAUUC--------- .....((((((..((....)).-----..))))))(((.((((...(((..........)))...))))(((((((((....))))))))).)))...--------- ( -28.10) >consensus CGAGUGCGCUAA_GUGAAG________AAGG__GCGGUGAAGAGGACUCC_ACCGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUU .((((.((((..............................)))).))))...........(((.((((.(((((((((....))))))))).)))))))........ (-20.68 = -21.04 + 0.36)

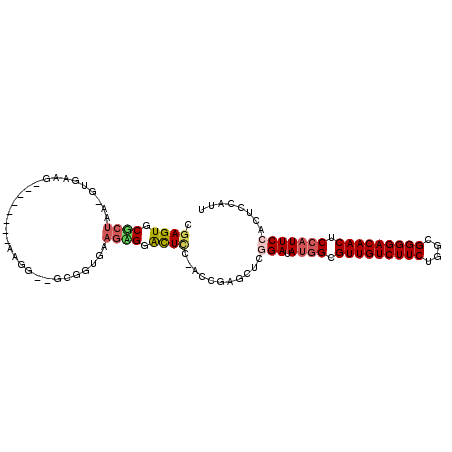
| Location | 16,663,560 – 16,663,673 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -21.58 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16663560 113 + 23771897 ACAUUGCUGGAGAAUGAGUCAUCCGCACUCGAAGAGUUCGCUGGCAGCAGCAGCAACAUUAGGUUGCGAAUCGCCCAGAUUCUUGCCGGCAAA-AAACAAGUACACAAAAAAAA ....(((((((((.....)).)))(.((((...)))).)((((((((.....(((((.....)))))(((((.....)))))))))))))...-.....))))........... ( -31.00) >DroSec_CAF1 31337 112 + 1 ACAUUGCUGGAGAAUGAGUCAUCCGCACUCGAAGAUUUCGCUGGCAACAGCAGCAGCAUUAGGUUGCGAAUCGCCCAGAUUCUUGCCGGCAAAAAAACAAGUACACAAA--AUA ...(((((((((((((((((....).))))...(((((.((((....)))).(((((.....))))))))))......)))))..))))))).................--... ( -34.00) >DroSim_CAF1 31393 112 + 1 ACAUUGCUGGAGAAUGAGUCAUCCGCACUCGAAGAUUUCGCUGGCAACAGCAGCAGCAUUAGGUUGCGAAUCGCCCAGAUUCUUGCCGGCAAAAAAACAAGUGCACAAA--AAA ........(((((.....)).)))(((((.((.....))((((((((.....(((((.....)))))(((((.....))))))))))))).........))))).....--... ( -34.30) >DroEre_CAF1 31465 102 + 1 ACAUUGCUGGAGAAUGAGUCAUCCGCACUCAAAGAGUUCUCUUGC---------AGCAUUAGGUUGCGAAUCGCCCAGAUUCUUGCCGGCAAA-AAACAAGUACACAAA--AAA ...(((((((....((((((....).)))))((((....))))((---------(((.....)))))(((((.....)))))...))))))).-...............--... ( -26.10) >DroYak_CAF1 32550 102 + 1 ACAUUGCUGGAGCAUGAGUCAUCCGCACUCAAAGAGUGCUCUUGC---------AGCAUUAGGUUGCGAAUCGCCCAGAUUCUUGCCGGCAAA-AAACAAGUACACAAA--AAA ...(((((((.....(((((....((((((...))))))..((((---------(((.....)))))))........)))))...))))))).-...............--... ( -31.90) >consensus ACAUUGCUGGAGAAUGAGUCAUCCGCACUCGAAGAGUUCGCUGGCA_CAGCAGCAGCAUUAGGUUGCGAAUCGCCCAGAUUCUUGCCGGCAAA_AAACAAGUACACAAA__AAA ...(((((((.(((((((((....).))))).....)))((((....)))).(((((.....)))))(((((.....)))))...)))))))...................... (-21.58 = -23.26 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:55 2006