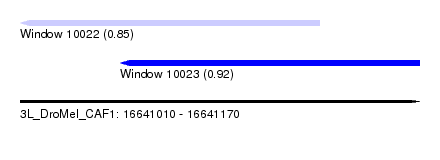
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,641,010 – 16,641,170 |
| Length | 160 |
| Max. P | 0.924194 |

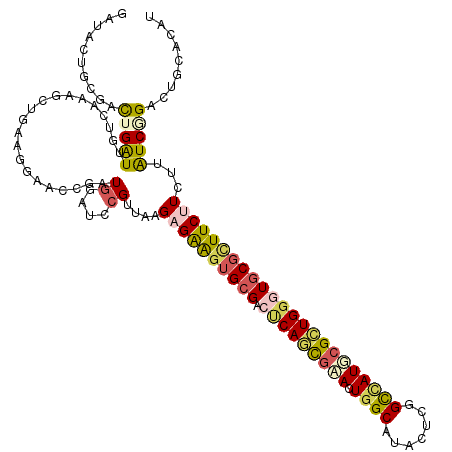
| Location | 16,641,010 – 16,641,130 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16641010 120 - 23771897 UCCGUUAAGAGAAGUGCGACUCAGCGAACUGGCGUAGUCGGCCAUUCGCUGGGUGCGUUUCUUCUUAUCGGACUGCACAUCGAGUUCCUUCUCCAGGUCCCUGAUCUUGACCUCCAGUCG ..((.((((((((((((.((((((((((.((((.......))))))))))))))))))))).))))).))(((((......(((......))).(((((.........))))).))))). ( -47.20) >DroVir_CAF1 11854 120 - 1 CCCGGUCAGUGGAGUGCGUCCCAGUGCAUUGGCGUACUCGGUUAUGCGCUGGCUGCGCUUCUUGCGAUCAGCCUGCACGUCCAGCUCCUUCUCCAGCUCACGUAUCUUGGCCUCCAUGCG ...((((((((((((((.....(((((((.(.((....)).).)))))))((((((((.....)))..))))).)))).))))(((........))).........))))))........ ( -42.10) >DroPse_CAF1 14151 120 - 1 UCCGUUAAGAGAAGAGCGACUCAGCGAACUGGCAUACUCGGCCAUUCGCUGGGUGCGCUUCUUCUUAUCGGACUGGACGUCCAGCUCCUUCUCCAUUUCCCGGAUCUUCGCCUCCAUGCG ((((....((((((((((((((((((((.((((.......)))))))))))))).))).)))))))...(((((((....)))).)))............))))....(((......))) ( -50.50) >DroGri_CAF1 12512 120 - 1 GCCGGUUAGUGGAAUGCGACCCACUUGAUUGGCAUAUUCGGUGAUGCGUUGACUGCGCUUCUUGCGAUCCGUCUGCACAUCCAGCUCCUUCUCCAGGUCACGUAUCUUCACCUCCAUGCG ((((((((((((........)))).))))))))......((((((((((.((((.(((.....)))....(.(((......))))..........)))))))))...)))))........ ( -36.20) >DroWil_CAF1 18103 120 - 1 ACCGCUCAGAGAGGUGCGACUCAAUGAAUUGGCAUAAUCGGCCAUGCGUUGGGUACGCUUCUUUUUAUCCGAUUGCACAUCCAGUUCCUUCUCCAACUCUCGAAUUUUCGCCUCCAUGCG ..(((...(.((((((((((((((((...((((.......))))..)))))))).)))............(((((......))))).......................))))))..))) ( -31.10) >DroPer_CAF1 12593 120 - 1 UCCGUUAAGAGAAGAGCGACUCAGCGAACUGGCAUACUCGGCCAUUCGCUGGGUGCGCUUCUUCUUAUCGGACUGGACGUCCAGCUCCUUCUCCAUUUCCCGGAUCUUCGCCUCCAUGCG ((((....((((((((((((((((((((.((((.......)))))))))))))).))).)))))))...(((((((....)))).)))............))))....(((......))) ( -50.50) >consensus UCCGUUAAGAGAAGUGCGACUCAGCGAACUGGCAUACUCGGCCAUGCGCUGGGUGCGCUUCUUCUUAUCGGACUGCACAUCCAGCUCCUUCUCCAGCUCCCGGAUCUUCGCCUCCAUGCG ........((((((.(((((((((((((.((((.......)))))))))))))).)))..............(((......)))...))))))........................... (-24.23 = -24.68 + 0.45)

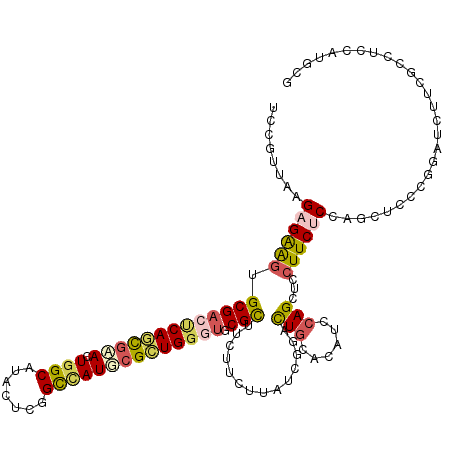
| Location | 16,641,050 – 16,641,170 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -44.78 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16641050 120 - 23771897 GGUACUGGGACUGAUUUUCGAAGCAGAAGGAACCAAUCGAUCCGUUAAGAGAAGUGCGACUCAGCGAACUGGCGUAGUCGGCCAUUCGCUGGGUGCGUUUCUUCUUAUCGGACUGCACAU ......................((((..(....)......((((.((((((((((((.((((((((((.((((.......))))))))))))))))))))).))))).)))))))).... ( -44.70) >DroVir_CAF1 11894 120 - 1 GGCACUGCGACUGAUUGUCAAAGCUAAACGAACUGAUGGACCCGGUCAGUGGAGUGCGUCCCAGUGCAUUGGCGUACUCGGUUAUGCGCUGGCUGCGCUUCUUGCGAUCAGCCUGCACGU ...((((((.(((((((.(((..........((((((.(...).))))))((((((((..(((((((((.(.((....)).).))))))))).))))))))))))))))))..)))).)) ( -47.20) >DroGri_CAF1 12552 120 - 1 GACACUGCGACUGAUUGUCAAAGCUGAAGGAGCUGAUGGAGCCGGUUAGUGGAAUGCGACCCACUUGAUUGGCAUAUUCGGUGAUGCGUUGACUGCGCUUCUUGCGAUCCGUCUGCACAU .....((((((.....)))..((((.....))))((((((((((((((((((........)))).))))))))....(((..((.((((.....)))).))...))))))))).)))... ( -40.90) >DroWil_CAF1 18143 120 - 1 GAUAUUGCGAUUGAUUCUCAAAACUAAAUGAACCAAUGGAACCGCUCAGAGAGGUGCGACUCAAUGAAUUGGCAUAAUCGGCCAUGCGUUGGGUACGCUUCUUUUUAUCCGAUUGCACAU .....((((((((....(((........)))................(((((((.(((((((((((...((((.......))))..)))))))).))).)))))))...))))))))... ( -34.50) >DroAna_CAF1 11460 120 - 1 GGUACUGGGACUGAUUCUCGAAGCUGAAGGAACUGAUGGAUCCGUUAAGAGAAGUGCGACUCAGCGAACUGGCAUAGUCGGCCAUUCGCUGGGUGCGCUUCUUCUUGUCGGACUGCACAU (((..(((((.....)))))..)))........((.(((.((((.((((((((((((.((((((((((.((((.......))))))))))))))))))))).))))).)))))))))... ( -48.70) >DroPer_CAF1 12633 120 - 1 GAUAGAGGGAUUGGUUGUCGAAAGAGAACGACCCGAUGGAUCCGUUAAGAGAAGAGCGACUCAGCGAACUGGCAUACUCGGCCAUUCGCUGGGUGCGCUUCUUCUUAUCGGACUGGACGU ..(((.(((..(.(((.((....)).))).))))......((((.(((((((((.((.((((((((((.((((.......)))))))))))))))).)))).))))).)))))))..... ( -52.70) >consensus GAUACUGCGACUGAUUGUCAAAGCUGAAGGAACCGAUGGAUCCGUUAAGAGAAGUGCGACUCAGCGAACUGGCAUACUCGGCCAUGCGCUGGGUGCGCUUCUUCUUAUCGGACUGCACAU ..........(((((.....................((....))....((((((((((.(((((((((.((((.......)))))))))))))))))))))))...)))))......... (-23.88 = -24.42 + 0.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:46 2006