| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,614,599 – 16,614,746 |
| Length | 147 |
| Max. P | 0.995281 |

| Location | 16,614,599 – 16,614,712 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -27.04 |
| Energy contribution | -28.85 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
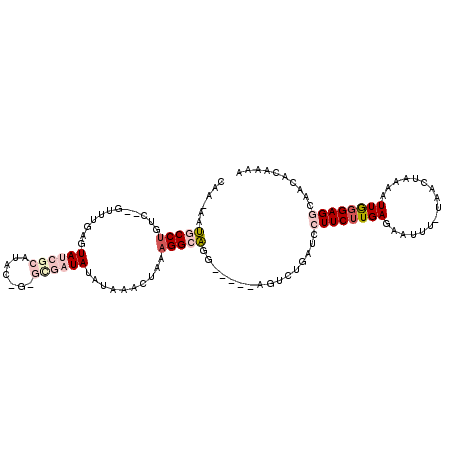
>3L_DroMel_CAF1 16614599 113 + 23771897 UAGGAAAACAGUUCUCCAGUAAAUCAAAUUCAAUUUCAACCAAAAAUGCCUGUCGUUCAAGUAUCGCAAACUGUGCGAUAUCCAAACUGAAGGCAGGAGUCUGAUCCUUCUUG .((((...(((..((((.(.((((........))))).........(((((...(((...((((((((.....))))))))...)))...))))))))).))).))))..... ( -28.70) >DroSim_CAF1 21880 113 + 1 UAGGAAAACAGUUCUCCAGCAAAUCAAAUUCAAUUUCAACCAAAAAUGCCUGUCGUUCGAGUAUCGCAUACUGUGUGAUAUCCAAACUGAAGGCAGGAGUCUGAUCCUUCUUG .((((...(((..((((.(.((((........))))).........(((((...(((.(..((((((((...))))))))..).)))...))))))))).))).))))..... ( -28.80) >DroEre_CAF1 22643 113 + 1 UUGGAAAACAGUUCUCCAGCAAAUCAAAUUCAAUUUCAACCAAGAUUGCCUGUCGUUUGAGUAUCGCAUACAGAGCGAUAUUUACACUAAAGGCAGGAGUCUGAUCCUUCUUG ..(((...(((..((((..................((......)).(((((...((.((((((((((.......)))))))))).))...))))))))).))).)))...... ( -31.20) >DroYak_CAF1 22822 113 + 1 CUGGAAAACAGUUUUCCAGCAAAUCAAAUUCUAUUUCAACCAAGAAUGCCCGACGUUUGAUUAUCGCAUACAAAGCGAUAUUGAAACUUUAGGCAGGAGUCUGAUCCUUCUUG (((((((.....))))))).....................((((((((((..(.((((.(.((((((.......)))))).).)))).)..))))(((......))))))))) ( -32.50) >consensus UAGGAAAACAGUUCUCCAGCAAAUCAAAUUCAAUUUCAACCAAAAAUGCCUGUCGUUCGAGUAUCGCAUACAGAGCGAUAUCCAAACUGAAGGCAGGAGUCUGAUCCUUCUUG ..(((...(((..((((.(.((((........))))).........(((((...(((((((((((((.......)))))))))))))...))))))))).))).)))...... (-27.04 = -28.85 + 1.81)

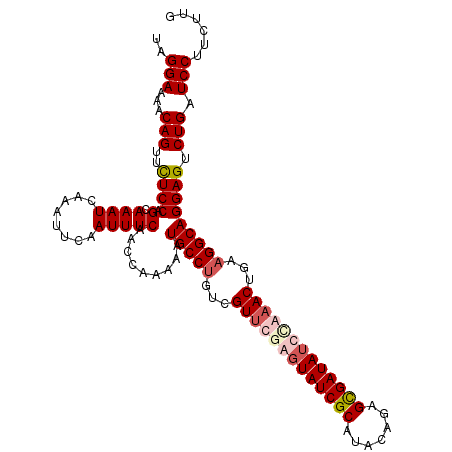
| Location | 16,614,599 – 16,614,712 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16614599 113 - 23771897 CAAGAAGGAUCAGACUCCUGCCUUCAGUUUGGAUAUCGCACAGUUUGCGAUACUUGAACGACAGGCAUUUUUGGUUGAAAUUGAAUUUGAUUUACUGGAGAACUGUUUUCCUA .....((((.(((.(((((((((...(((..(.(((((((.....))))))).)..)))...))))).....(((..((......))..)))....))))..)))...)))). ( -34.00) >DroSim_CAF1 21880 113 - 1 CAAGAAGGAUCAGACUCCUGCCUUCAGUUUGGAUAUCACACAGUAUGCGAUACUCGAACGACAGGCAUUUUUGGUUGAAAUUGAAUUUGAUUUGCUGGAGAACUGUUUUCCUA .....((((.(((.(((((((((...(((((..((((.((.....)).))))..)))))...))))).....(((..(((((......))))))))))))..)))...)))). ( -31.20) >DroEre_CAF1 22643 113 - 1 CAAGAAGGAUCAGACUCCUGCCUUUAGUGUAAAUAUCGCUCUGUAUGCGAUACUCAAACGACAGGCAAUCUUGGUUGAAAUUGAAUUUGAUUUGCUGGAGAACUGUUUUCCAA ......(((.(((.(((((((((...((.....((((((.......)))))).....))...))))).....(((..(((((......))))))))))))..)))...))).. ( -28.30) >DroYak_CAF1 22822 113 - 1 CAAGAAGGAUCAGACUCCUGCCUAAAGUUUCAAUAUCGCUUUGUAUGCGAUAAUCAAACGUCGGGCAUUCUUGGUUGAAAUAGAAUUUGAUUUGCUGGAAAACUGUUUUCCAG (((((((((......)))(((((...((((.(.((((((.......)))))).).))))...)))))))))))((..((......))..))...(((((((.....))))))) ( -31.20) >consensus CAAGAAGGAUCAGACUCCUGCCUUCAGUUUGAAUAUCGCACAGUAUGCGAUACUCAAACGACAGGCAUUCUUGGUUGAAAUUGAAUUUGAUUUGCUGGAGAACUGUUUUCCAA ......(((.(((.(((((((((...((((((.((((((.......)))))).))))))...))))).....(((..((......))..)))....))))..)))...))).. (-27.67 = -28.18 + 0.50)

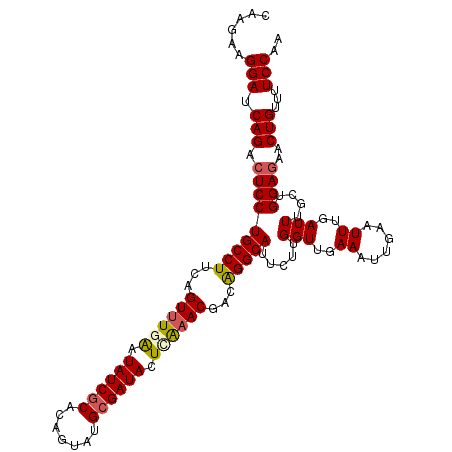

| Location | 16,614,639 – 16,614,746 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.49 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -8.39 |
| Energy contribution | -9.97 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16614639 107 + 23771897 CAAAAAUGCCUGUC--GUUCAAGUAUCGCAAACUGUGCGAUAUCCAAACUGAAGGCAGG-----AGUCUGAUCCUUCUUGAGAAUUU-UAACUAAAAUUGGGAGGCAACACAAAA ......(((((.((--(.....((((((((.....)))))))).....((.((((.(((-----(......)))))))).)).....-..........))).)))))........ ( -28.80) >DroPse_CAF1 24259 99 + 1 ------CGCCUAUUCUGCCUGAAUAUCU-AUAU---CCCUUAGAUACACUAGAGGGGGGUACAGCGUGU----GUUCUUGAGAAUUUCUAACUA-ACUUAGGAGGCAACACAA-A ------.(((((((((.(..((((((((-.(((---(((((((.....)))...))))))).))...))----))))..)))))).(((((...-..))))))))).......-. ( -23.50) >DroSim_CAF1 21920 107 + 1 CAAAAAUGCCUGUC--GUUCGAGUAUCGCAUACUGUGUGAUAUCCAAACUGAAGGCAGG-----AGUCUGAUCCUUCUUGAGAAUUU-UAACUAAAAUUGGGAGGCAACACAAAA ......(((((.((--(...(..((((((((...))))))))..)...((.((((.(((-----(......)))))))).)).....-..........))).)))))........ ( -28.70) >DroEre_CAF1 22683 107 + 1 CAAGAUUGCCUGUC--GUUUGAGUAUCGCAUACAGAGCGAUAUUUACACUAAAGGCAGG-----AGUCUGAUCCUUCUUGAGAAUUU-UAACUAAAAUUGGGAGGCAACACAAAA ..(((((.((((((--((.((((((((((.......)))))))))).))....))))))-----)))))...((((((....(((((-(....)))))))))))).......... ( -30.30) >DroYak_CAF1 22862 107 + 1 CAAGAAUGCCCGAC--GUUUGAUUAUCGCAUACAAAGCGAUAUUGAAACUUUAGGCAGG-----AGUCUGAUCCUUCUUGAGAAUUU-UAACUAAAAUUGGGAGGCAACACAAAA ((((((((((..(.--((((.(.((((((.......)))))).).)))).)..))))((-----(......)))))))))..(((((-(....))))))....(....)...... ( -27.90) >DroPer_CAF1 35145 99 + 1 ------CGCCUAUUCUGCCUGAAUAUCU-AUAU---CCCUUAGAUACACUAGAGGGGGGUACAGCGUGU----GUUCAUGAGAAUUUCUAACUA-ACUUAGGAGGCAACACAA-A ------.(((((((((.(.(((((((((-.(((---(((((((.....)))...))))))).))...))----))))).)))))).(((((...-..))))))))).......-. ( -25.00) >consensus CAA_AAUGCCUGUC__GUUUGAGUAUCGCAUAC_G_GCGAUAUAUAAACUAAAGGCAGG_____AGUCUGAUCCUUCUUGAGAAUUU_UAACUAAAAUUGGGAGGCAACACAAAA ......(((((............((((((.......))))))..........)))))................((((((((................)))))))).......... ( -8.39 = -9.97 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:40 2006