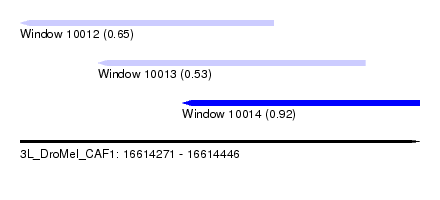
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,614,271 – 16,614,446 |
| Length | 175 |
| Max. P | 0.918296 |

| Location | 16,614,271 – 16,614,382 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16614271 111 - 23771897 UAACAGUCGAAAAGACGUCUGAUAAAAUGUUGCUGACUGACUGGAUUAAGUCGA---AGCAUUGUCAUAGUCACCAAUAAAAUAUUUUUGAA-AUUUUAACA-----UAUAUUCCAGUCA .....(((.....)))(.(((......(((((.(((((((((......))))((---.......))..))))).))))).(((((..((((.-...))))..-----.))))).))).). ( -18.70) >DroSec_CAF1 22319 114 - 1 UAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGAAGAAGCAUUGUCGUAGUCACCAAUAAAUUAUUUUUGAAAAUUGUAACA-----UAUUUGGCCGUCA .....(((...-.)))(((.(.(((((((((((....(((((((((.....(......)....))).)))))).(((..........)))......))))))-----).)))).).))). ( -22.40) >DroSim_CAF1 21550 114 - 1 UAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGAAGAAGCAUUGUCGUAGUCACCAAUAAAUUAUUUUUGAAAAUUGUAACA-----UAUUUGCCAGUCA .....(((...-.)))(((((.(((((((((((....(((((((((.....(......)....))).)))))).(((..........)))......))))))-----).)))).))))). ( -27.00) >DroEre_CAF1 22308 112 - 1 UAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGA---AGCAUUGUCAUAGUCACCAAUAAAAUAUUUUUGAAAAUU-UAACA---UAUAUAUUCCAGUCA .....(((...-.)))(((((......(((((.((((((((.............---......)))..))))).))))).(((((...((......-...))---...))))).))))). ( -22.61) >DroYak_CAF1 22484 115 - 1 UAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGACUAAAUUAA---AGCAUUGUCAUAGUCACCAAUACAAUAUUUUUGAAAAUU-UAAAAAUGUAUAUAUUCCAGUCA .....(((...-.)))(((((......(((((.((((((....(((........---......))).)))))).)))))..((((((((((....)-)))))))))........))))). ( -29.54) >consensus UAACAGUCGAA_AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGA___AGCAUUGUCAUAGUCACCAAUAAAAUAUUUUUGAAAAUU_UAACA_____UAUAUUCCAGUCA ...((((((......))))))......(((((.((((((....(((.................))).)))))).)))))......................................... (-17.21 = -17.25 + 0.04)

| Location | 16,614,305 – 16,614,422 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16614305 117 - 23771897 AAUAAACUGUUUUCAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAAAAGACGUCUGAUAAAAUGUUGCUGACUGACUGGAUUAAGUCGA---AGCAUUGUCAUAGUCACCAAUAA .....((((((........(((........)))........))))))......(((...((((((.....((((...(((((......))))).---))))))))))..)))........ ( -20.99) >DroSec_CAF1 22354 119 - 1 AAUAAACUGUUUUCAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGAAGAAGCAUUGUCGUAGUCACCAAUAA .......((((((..........))))))..((((((((.((.((((((..-...)))))).)))))))))).....(((((((((.....(......)....))).))))))....... ( -20.30) >DroSim_CAF1 21585 119 - 1 AAUAAACUGUUUUCAAAUCUGUAAGAAAAAACAAUAUUUAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGAAGAAGCAUUGUCGUAGUCACCAAUAA .........(((((..........)))))..((((((((.((.((((((..-...)))))).)))))))))).....(((((((((.....(......)....))).))))))....... ( -19.70) >DroEre_CAF1 22344 116 - 1 AAUAAACUGUUUUGAAAUCUGUAAGAACAAACCAUAUUGAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGA---AGCAUUGUCAUAGUCACCAAUAA .......((((((..........)))))).....(((((......(((...-.)))(((((....((((((..(((..............))).---))))))....)))))..))))). ( -20.54) >DroYak_CAF1 22523 116 - 1 AAUAAACUGUUUUGAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGACUGGACUAAAUUAA---AGCAUUGUCAUAGUCACCAAUAC .......((((((..........))))))..((((((((.((.((((((..-...)))))).)))))))))).....(((((((((........---......))).))))))....... ( -23.54) >consensus AAUAAACUGUUUUCAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAA_AGACGACUGAUAAAAUGUUGCUGACUGACUGGAUUAAAUCGA___AGCAUUGUCAUAGUCACCAAUAA .......((((((..........))))))..((((((((.((.((((((......)))))).)))))))))).....(((((((((.................))).))))))....... (-17.83 = -18.47 + 0.64)


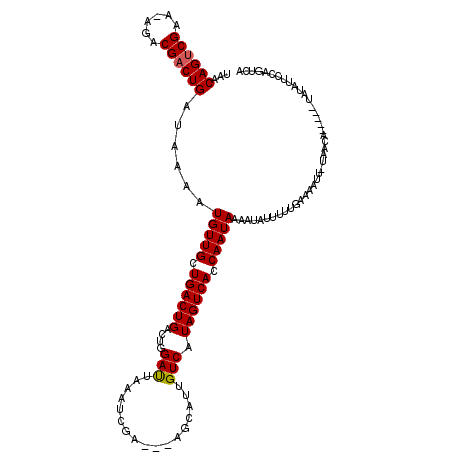
| Location | 16,614,342 – 16,614,446 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 97.10 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16614342 104 - 23771897 GAGCGCAAAUUGUUAUUGUAAAGUAAUAAACUGUUUUCAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAAAAGACGUCUGAUAAAAUGUUGCUGACUGA (((.(((..((((((((....))))))))..))).))).................((((((((.((.(((.((......)).))).))))))))))........ ( -17.80) >DroSec_CAF1 22394 103 - 1 GAGCGCAAAUUGUUAUUGUAAAAUAAUAAACUGUUUUCAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGA (((.(((..((((((((....))))))))..))).))).................((((((((.((.((((((..-...)))))).))))))))))........ ( -21.50) >DroSim_CAF1 21625 103 - 1 GAGCGCAAAUUGUUAUUGUAAAGUAAUAAACUGUUUUCAAAUCUGUAAGAAAAAACAAUAUUUAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGA (((.(((..((((((((....))))))))..))).))).................((((((((.((.((((((..-...)))))).))))))))))........ ( -21.50) >DroEre_CAF1 22381 103 - 1 GAGCGCAAAUUGUUAUUGUAAAGUAAUAAACUGUUUUGAAAUCUGUAAGAACAAACCAUAUUGAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGA .((((((..((((((((....))))))))..((((((..........))))))..............((((((..-...))))))......)).))))...... ( -18.50) >DroYak_CAF1 22560 103 - 1 GAGCGCAAAUUGUUAUUGUAAAGUAAUAAACUGUUUUGAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAA-AGACGACUGAUAAAAUGUUGCUGACUGA .(((.....((((((((....))))))))..((((((..........))))))...(((((((.((.((((((..-...)))))).))))))))))))...... ( -19.80) >consensus GAGCGCAAAUUGUUAUUGUAAAGUAAUAAACUGUUUUCAAAUCUGUAAGAACAAACAAUAUUUAUAACAGUCGAA_AGACGACUGAUAAAAUGUUGCUGACUGA .((((((..((((((((....))))))))..)))......................(((((((.((.((((((......)))))).))))))))))))...... (-17.20 = -17.64 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:36 2006