
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,605,473 – 16,605,616 |
| Length | 143 |
| Max. P | 0.940783 |

| Location | 16,605,473 – 16,605,586 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
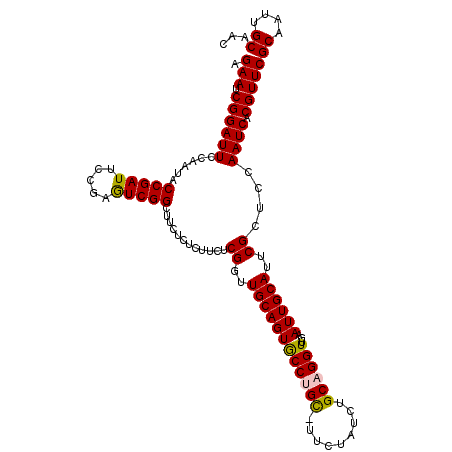
>3L_DroMel_CAF1 16605473 113 - 23771897 AGAAUCGGAUUCCAAUACCGAUUUCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCUGCCUUCUAUCUGCAGGUUGAAUUGCAUUCGCUCCAAUCACGUUCGCAAUUGCAAC .(((((((.........)))))))((((..((......))..))))(((((...((((((.........))))))..((((((...((.........))...))))))))))) ( -28.30) >DroSec_CAF1 13503 112 - 1 AGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCAGC-UUCUAUCUGCAGGUUGAAUUGCAUUCGCUCCAAUCACGUUCGCAAUUGCAAC .(((((((.........)))))))((((..((......))..))))((((((((((..((-........)).(((((....((....))..)))))......)).)))))))) ( -30.00) >DroSim_CAF1 12694 112 - 1 AGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCAGC-UUCUAUCUGCAGGUUGAAUUGCAUUCGCUCCAAUCACGUUCGCAAUUGCAAC .(((((((.........)))))))((((..((......))..))))((((((((((..((-........)).(((((....((....))..)))))......)).)))))))) ( -30.00) >DroEre_CAF1 13519 112 - 1 AGAAUCGGAUUCCGAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUACCUGU-CUCUAUCUGCAGGUUGAAUUGCAUUCGCUCCAAUCACGUUCGCAAUUGCAAC .(((.((((..(((((((.......)))))))..))........((..((((((((((((-........))))))...))))))..)).........)))))((....))... ( -30.00) >DroYak_CAF1 13558 112 - 1 AGAAUCGGAUUCCAAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUACCUGU-CUCUAUCUGCAGGUUGGAUUGCAUUCGCUCCAAUCACGUUCGCAAUUGCAAC .((((((((.......)))))))).(((.((.................(((((.......-......)))))(((((((..((....))))))))).)))))((....))... ( -30.42) >consensus AGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCUGC_UUCUAUCUGCAGGUUGAAUUGCAUUCGCUCCAAUCACGUUCGCAAUUGCAAC .(((.((((((......(((((.....)))))............((..((((((((((((.........))))))...))))))..))....)))).)))))((....))... (-25.36 = -25.04 + -0.32)

| Location | 16,605,507 – 16,605,616 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -32.71 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.54 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16605507 109 - 23771897 GAAUCCAUCAUUGAGCCAUGAGAA------GUAUAAAGAAUCGGAUUCCAAUACCGAUUUCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCUGCCUUCUAUCUGCAGGUUGAA ........(((((((((..(((((------(.....((((.(.(((((.((.......)).))))).).))))..)))))))))).)))))(((((.........)))))..... ( -28.70) >DroSec_CAF1 13537 108 - 1 GAAUCCAUCAUUGAGCCAUGAGAA------GUAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCAGC-UUCUAUCUGCAGGUUGAA ...((.(((((((((....(((((------((.....((.(((((.((.......)).))))).)).)))))))....))))))(((((.......-......)))))))).)). ( -30.42) >DroSim_CAF1 12728 108 - 1 GAAUCCAUCAUUGAGCCAUGAGAA------GUAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCAGC-UUCUAUCUGCAGGUUGAA ...((.(((((((((....(((((------((.....((.(((((.((.......)).))))).)).)))))))....))))))(((((.......-......)))))))).)). ( -30.42) >DroEre_CAF1 13553 108 - 1 GAAUCCAUCACUGAGCCAUGAGAA------GUAUAAAGAAUCGGAUUCCGAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUACCUGU-CUCUAUCUGCAGGUUGAA .......((((((....(((((((------((.....(((((((...))))))).((..(((...)))..))..)))))))))...))))((((((-........)))))).)). ( -32.80) >DroYak_CAF1 13592 114 - 1 GAAUCCAUCAUUGAGCCAUGAGAAGUAGAAGUAUAAAGAAUCGGAUUCCAAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUACCUGU-CUCUAUCUGCAGGUUGGA ...((((.......((.(((((((((((((((.....((((((((.......)))))))).......)))))).)))))))))..))...((((((-........)))))))))) ( -41.20) >consensus GAAUCCAUCAUUGAGCCAUGAGAA______GUAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCUGC_UUCUAUCUGCAGGUUGAA ...........(.((((.((((((......((.....((.(((((.((.......)).))))).)).)).......))))))..(((((..............))))))))).). (-22.86 = -22.54 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:30 2006