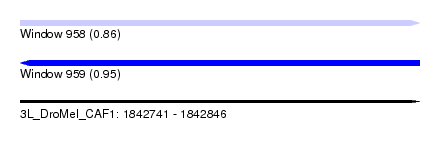
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,842,741 – 1,842,846 |
| Length | 105 |
| Max. P | 0.951549 |

| Location | 1,842,741 – 1,842,846 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
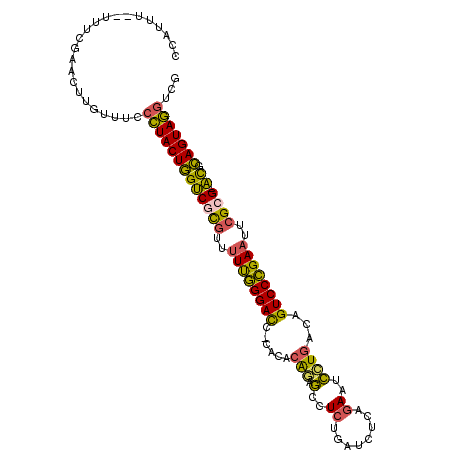
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -22.99 |
| Energy contribution | -22.47 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
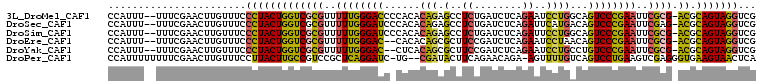
>3L_DroMel_CAF1 1842741 105 + 23771897 CGACCUACUGCGU-CGCGAAUUCGGGACUGCCAGGAUUCUGAGAUCAGAGGCUCUGUGUGGGGUCCCAAAAACGCGACCAGUAGGGAAACAAGUUCGAAA--AAAUGG ...(((((((.((-((((..((.((((((((((((.(((((....)))))..)))).))..)))))).))..)))))))))))))...............--...... ( -43.70) >DroSec_CAF1 1952 105 + 1 CGACCUACUGCGU-CUCGAAUUCGGGACUGUCAUGAAUCUGAGAUCAGAGGCUCUGUGUGGGAUCCCAAAAACGCGACCAGUAGGGAAACAAGUUCGAAA--AAAUGG ...(((((((.((-((((....)))))).(((..(..((((....))))..)...((.(((....)))...))..))))))))))...............--...... ( -31.90) >DroSim_CAF1 1957 105 + 1 CGACCUACUGCGU-CGCGAAUUCGGGACUGCCAGGAAUCUGAGAUCAGAGGCUCUGUGUGGGAUCCCAAAAACGCGACCAGUAGGGAAACAAGUUCGAAA--AAAUGG ...(((((((.((-((((..((.(((((..(((((..((((....))))..).))).)..)..)))).))..)))))))))))))...............--...... ( -41.80) >DroEre_CAF1 1945 103 + 1 CGACCUACUGCGU-CGCGAAUUCGGGACUGUUAGGAUUCUGAGAUCGGAAGCGCUGUGUG--GUCCCAAAAACGCGACCAGUAGGGAAACAAGUUCGAAA--AAAUGG ...(((((((.((-((((..((.(((((..(((((.(((((....))))).).))).)..--))))).))..)))))))))))))...............--...... ( -40.70) >DroYak_CAF1 1952 103 + 1 CGACCUACUGCGU-CGCGAAUUCGGGACAGGCAGGAUUCUGAGAUCGGAAGCGCUGUGAG--GUCCCAAAAACGCGACCAGUAGGGAAACAAGUUCGAAA--AAAUGG ...(((((((.((-((((..((.(((((..(((((.(((((....))))).).))))...--))))).))..)))))))))))))...............--...... ( -42.60) >DroPer_CAF1 2022 104 + 1 UGAGUUACUUCACCCUCGACUUCAGGACUGACAAAACU-UCUGUUCUGAAGUAUCG--CA-GAUCCUGAGCGGACGGCAAGUAAGGAAACAAGUUCGAAAAAAAAUGG .(((((((((..((.((...((((((((((.....(((-((......)))))....--))-).)))))))..)).)).))))))(....)...)))............ ( -25.80) >consensus CGACCUACUGCGU_CGCGAAUUCGGGACUGCCAGGAUUCUGAGAUCAGAAGCUCUGUGUG_GAUCCCAAAAACGCGACCAGUAGGGAAACAAGUUCGAAA__AAAUGG (((.((((((.((.((((..((.((((....((((..((((....))))..).))).......)))).))..))))))))))))(....)....)))........... (-22.99 = -22.47 + -0.52)

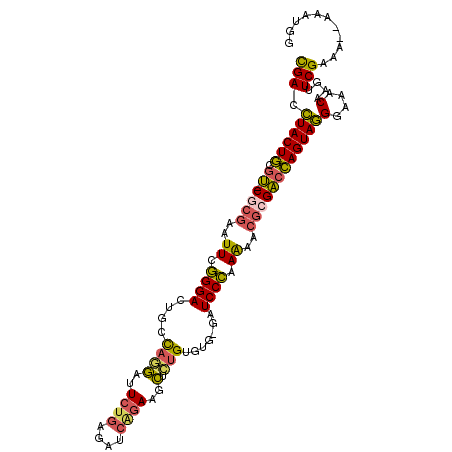
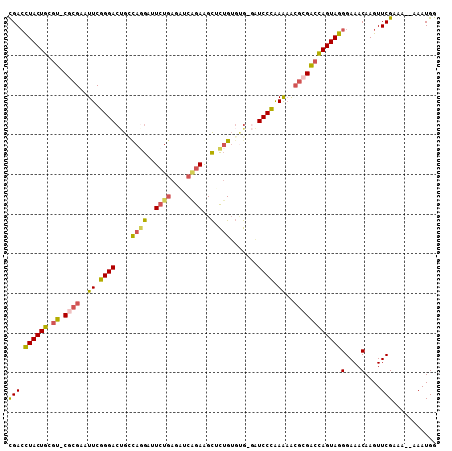
| Location | 1,842,741 – 1,842,846 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -27.17 |
| Energy contribution | -26.10 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1842741 105 - 23771897 CCAUUU--UUUCGAACUUGUUUCCCUACUGGUCGCGUUUUUGGGACCCCACACAGAGCCUCUGAUCUCAGAAUCCUGGCAGUCCCGAAUUCGCG-ACGCAGUAGGUCG ......--...............(((((((((((((..((((((((......(((.(..((((....))))..))))...))))))))..))))-)).)))))))... ( -43.30) >DroSec_CAF1 1952 105 - 1 CCAUUU--UUUCGAACUUGUUUCCCUACUGGUCGCGUUUUUGGGAUCCCACACAGAGCCUCUGAUCUCAGAUUCAUGACAGUCCCGAAUUCGAG-ACGCAGUAGGUCG ......--...............((((((((((.((..((((((((....((..(((..((((....))))))).))...))))))))..)).)-)).)))))))... ( -32.70) >DroSim_CAF1 1957 105 - 1 CCAUUU--UUUCGAACUUGUUUCCCUACUGGUCGCGUUUUUGGGAUCCCACACAGAGCCUCUGAUCUCAGAUUCCUGGCAGUCCCGAAUUCGCG-ACGCAGUAGGUCG ......--...............(((((((((((((..((((((((......(((.(..((((....))))..))))...))))))))..))))-)).)))))))... ( -42.00) >DroEre_CAF1 1945 103 - 1 CCAUUU--UUUCGAACUUGUUUCCCUACUGGUCGCGUUUUUGGGAC--CACACAGCGCUUCCGAUCUCAGAAUCCUAACAGUCCCGAAUUCGCG-ACGCAGUAGGUCG ......--...............(((((((((((((..((((((((--.....((.(.(((.(....).))).)))....))))))))..))))-)).)))))))... ( -37.40) >DroYak_CAF1 1952 103 - 1 CCAUUU--UUUCGAACUUGUUUCCCUACUGGUCGCGUUUUUGGGAC--CUCACAGCGCUUCCGAUCUCAGAAUCCUGCCUGUCCCGAAUUCGCG-ACGCAGUAGGUCG ......--...............(((((((((((((..((((((((--....(((.(.(((.(....).))).))))...))))))))..))))-)).)))))))... ( -40.40) >DroPer_CAF1 2022 104 - 1 CCAUUUUUUUUCGAACUUGUUUCCUUACUUGCCGUCCGCUCAGGAUC-UG--CGAUACUUCAGAACAGA-AGUUUUGUCAGUCCUGAAGUCGAGGGUGAAGUAACUCA ........................(((((((((.((.((((((((.(-((--(((.(((((......))-))).))).)))))))).))).)).))).)))))).... ( -33.00) >consensus CCAUUU__UUUCGAACUUGUUUCCCUACUGGUCGCGUUUUUGGGACC_CACACAGAGCCUCUGAUCUCAGAAUCCUGACAGUCCCGAAUUCGCG_ACGCAGUAGGUCG .......................(((((((((((((..((((((((......(((.(..((........))..))))...))))))))..)))).)).)))))))... (-27.17 = -26.10 + -1.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:01 2006