
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,543,683 – 16,543,792 |
| Length | 109 |
| Max. P | 0.779778 |

| Location | 16,543,683 – 16,543,792 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -19.23 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
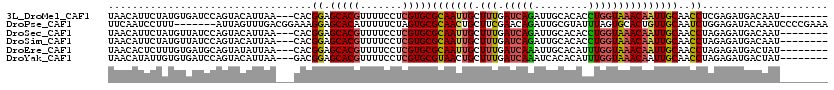
>3L_DroMel_CAF1 16543683 109 + 23771897 --------AUUGUCAUCUCGAGGUUGCAAUUGUUUACCAGGUGUGCAAUCUGAUCAAAGCAAUUGCGCACGAGGAAAACGUGCUCCGUG---UUAAUGUACUGGAUCACAUAGAAUGUUA --------....((.(((((..((.((((((((((..(((((.....)))))....)))))))))))).))))).....(((.((((.(---(......)))))).)))...))...... ( -30.10) >DroPse_CAF1 33989 113 + 1 UUUCGGGGAUUUGUAUCUCCAGAUUGCAACAGUGCACUAAAUACGCAAUCUGUUCGAAGCAGUUGCGCACUAGAAAAAUGUGCUCCUUUCCGUCAAACUAAU-------AAAGGAUUGAA ....((((((....))))))....((((....))))........((((.((((.....))))))))((((.........))))((((((.............-------))))))..... ( -26.42) >DroSec_CAF1 3808 109 + 1 --------AUUGUCAUCUCUAGGUUGCAAUUGUUUACCAGGUGUGCAAUCUGAUCAAAGCAAUUGCGCACGAGGAAAACGUGCUCCGUG---UUAAUGUACUGGAUAACAUAGAAUGUUA --------.....(((.((((.(((((((((((((..(((((.....)))))....))))))))))(((((.......)))))((((.(---(......)))))).))).)))))))... ( -29.60) >DroSim_CAF1 4375 109 + 1 --------AUUGUCAUCUCUAGGUUGCAAUUGUUUACCAGGUGUGCAAUCUGAUCAAAGCAAUUGCGCACGAGGAAAACGUGCUCCGUG---UUAAUGUACUGGAUAACAUAGAAUGUUA --------.....(((.((((.(((((((((((((..(((((.....)))))....))))))))))(((((.......)))))((((.(---(......)))))).))).)))))))... ( -29.60) >DroEre_CAF1 3822 109 + 1 --------AUAGUCAUCUCUAGGUUGCAAUUGUUUACCAAAUGUGCAAUUUGAUCAAAGCAAUUGCGCACGAGGAAAACGUGCUCCGUG---UUAAUAUACUGCAUCACAAAGAGUGUUA --------.....((.((((.((..((((((((((..(((((.....)))))....))))))))))(((((.......))))).))(((---(....))))..........))))))... ( -26.40) >DroYak_CAF1 3851 109 + 1 --------AUAGUCAUCUCUAGGUUGCAAUUGUUUACCAAAUGUGUGAUUUGAUCAAAGCAGUUACGCACGAGGAAAACGUGCUCCGUC---UUAAUGUACUGGAUCACACAAUAUGUUA --------.(((......)))(((.((....))..)))...(((((((..................(((((.......)))))((((((---.....).)).))))))))))........ ( -21.50) >consensus ________AUUGUCAUCUCUAGGUUGCAAUUGUUUACCAAAUGUGCAAUCUGAUCAAAGCAAUUGCGCACGAGGAAAACGUGCUCCGUG___UUAAUGUACUGGAUCACAUAGAAUGUUA .................(((((...((((((((((..(((((.....)))))....))))))))))(((((.......))))).................)))))............... (-19.23 = -20.15 + 0.92)



| Location | 16,543,683 – 16,543,792 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -17.68 |
| Energy contribution | -17.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
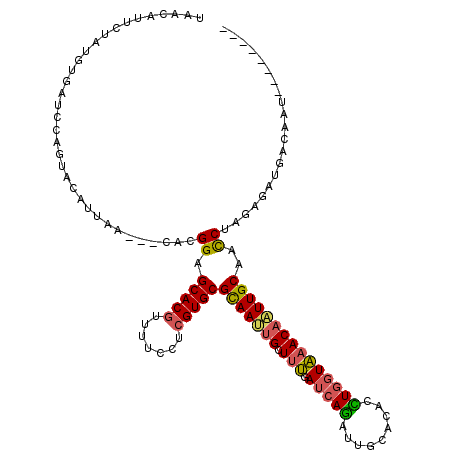
>3L_DroMel_CAF1 16543683 109 - 23771897 UAACAUUCUAUGUGAUCCAGUACAUUAA---CACGGAGCACGUUUUCCUCGUGCGCAAUUGCUUUGAUCAGAUUGCACACCUGGUAAACAAUUGCAACCUCGAGAUGACAAU-------- ..........(((.((((.((......)---)..((.(((((.......)))))(((((((.(((.(((((.........)))))))))))))))..))..).))).)))..-------- ( -26.70) >DroPse_CAF1 33989 113 - 1 UUCAAUCCUUU-------AUUAGUUUGACGGAAAGGAGCACAUUUUUCUAGUGCGCAACUGCUUCGAACAGAUUGCGUAUUUAGUGCACUGUUGCAAUCUGGAGAUACAAAUCCCCGAAA (((..(((.((-------(......))).)))..(((((.((((.....)))).))...((..((...(((((((((....(((....))).)))))))))..))..))..)))..))). ( -27.50) >DroSec_CAF1 3808 109 - 1 UAACAUUCUAUGUUAUCCAGUACAUUAA---CACGGAGCACGUUUUCCUCGUGCGCAAUUGCUUUGAUCAGAUUGCACACCUGGUAAACAAUUGCAACCUAGAGAUGACAAU-------- ..........((((((((.((......)---)..((.(((((.......)))))(((((((.(((.(((((.........)))))))))))))))..))..).)))))))..-------- ( -28.50) >DroSim_CAF1 4375 109 - 1 UAACAUUCUAUGUUAUCCAGUACAUUAA---CACGGAGCACGUUUUCCUCGUGCGCAAUUGCUUUGAUCAGAUUGCACACCUGGUAAACAAUUGCAACCUAGAGAUGACAAU-------- ..........((((((((.((......)---)..((.(((((.......)))))(((((((.(((.(((((.........)))))))))))))))..))..).)))))))..-------- ( -28.50) >DroEre_CAF1 3822 109 - 1 UAACACUCUUUGUGAUGCAGUAUAUUAA---CACGGAGCACGUUUUCCUCGUGCGCAAUUGCUUUGAUCAAAUUGCACAUUUGGUAAACAAUUGCAACCUAGAGAUGACUAU-------- ...((((((..(((.((........)).---)))((.(((((.......)))))(((((((.(((.(((((((.....)))))))))))))))))..)).)))).)).....-------- ( -26.40) >DroYak_CAF1 3851 109 - 1 UAACAUAUUGUGUGAUCCAGUACAUUAA---GACGGAGCACGUUUUCCUCGUGCGUAACUGCUUUGAUCAAAUCACACAUUUGGUAAACAAUUGCAACCUAGAGAUGACUAU-------- ...((...((((((((.......(((((---..(((.(((((.......)))))....)))..)))))...))))))))..))((((....)))).................-------- ( -21.60) >consensus UAACAUUCUAUGUGAUCCAGUACAUUAA___CACGGAGCACGUUUUCCUCGUGCGCAAUUGCUUUGAUCAGAUUGCACACCUGGUAAACAAUUGCAACCUAGAGAUGACAAU________ ..................................((.(((((.......)))))(((((((.(((.(((((.........)))))))))))))))..))..................... (-17.68 = -17.43 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:13 2006